sophieRAIBAUD
sophieRAIBAUD
hello I mean 90° turn in order to see the pictures (and the data) in the same orientation Many thanks !!!
OK thanks ! ill try it for the image but I also will need to rotate the spatial data
hello Giovanni, do you think I need to turn the spatial transcriptomic data in order to perform batch correction with MNN on the 2 data than are not spatially aligned...
ah OK thanks So I rotated -adata.obsm["spatial"] -adata.uns["spatial"] ["xxx"]["image"][....] and the results is really strange I lost all the picture (it's completly zoomed on 1 corner) I'll get back to...
Hello Giovanni, I have performed MNN on my 2 samples following this tutorial: https://nbisweden.github.io/workshop-scRNAseq/labs/compiled/scanpy/scanpy_03_integration.html on the PCA, tsne, and umap graphes, it look pretty cool: 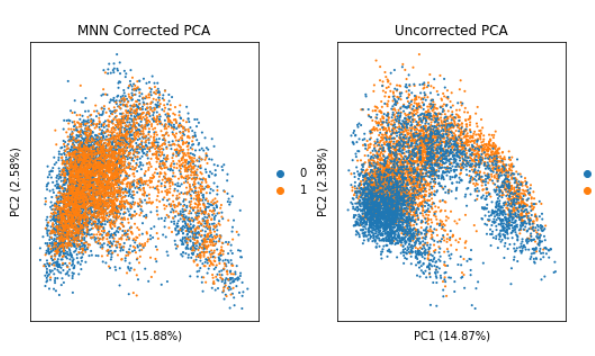 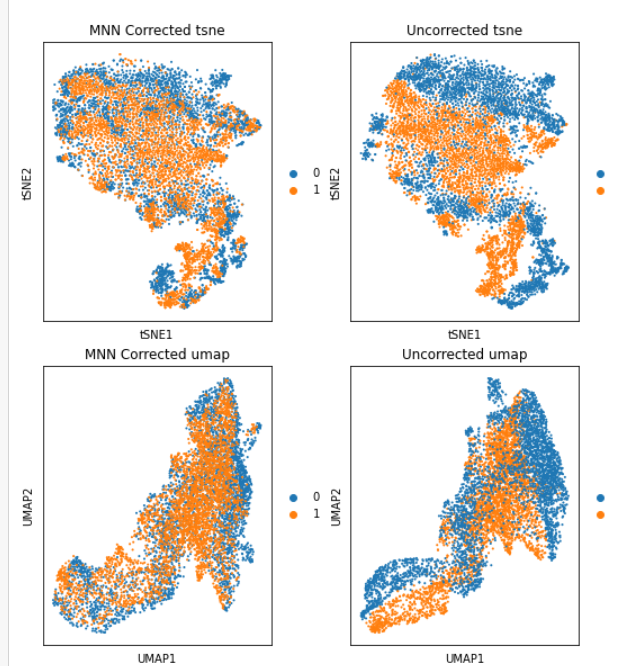 but now I...
I found the solution with this notebook https://nbisweden.github.io/workshop-scRNAseq/labs/compiled/scanpy/scanpy_07_spatial.html but I had to get back the image data: corrected.uns['spatial'] = adata_raw.uns['spatial'] it works !
> @sophieRAIBAUD I believe you don't need to rotate the image for the MNN. I'd be curious to see whether the image and coordiantes before rotation are nonetheless aligned? @giovp...
hello just want to analysis these kind of data with squidpy (https://www.akoyabio.com/) which functions work on codex data (I believe its .mav format) Thanks !!
@giovp many thanks, I'll try it asap
the first example is working properly thanks ! _import napari import squidpy as sq adata = sq.datasets.visium_hne_adata() img1 = adata.uns["spatial"]["V1_Adult_Mouse_Brain"]["images"]["hires"].copy() viewer = napari.Viewer() viewer.add_image( img1, rgb=True, name="image1", metadata={"adata": adata, "library_id":...