MNN correction on spatial datasets
Description
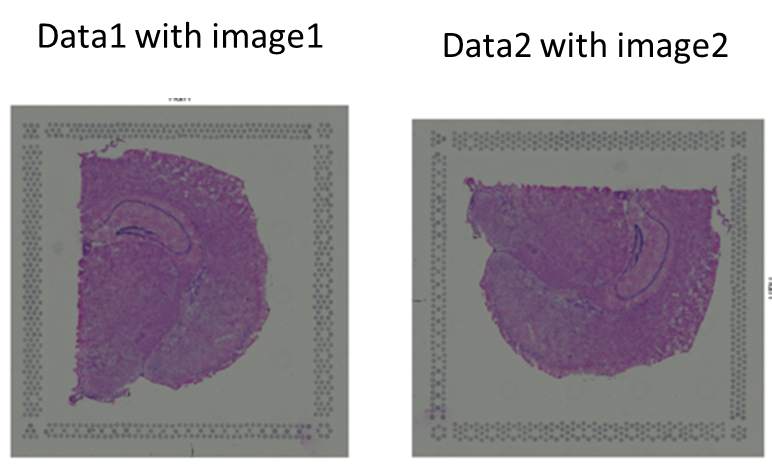
...hello I am struggling with 2 spatial transcriptomics data to compare, but the 2 tissues were spoted perpendicular to each other I dont know how to pivot the whole anndata (with image + transcripomic data) any idea ? Many thanks for this wonderfull package !!!
hi @sophieRAIBAUD thanks for the interest, what do you mean "pivot" ?
hello I mean 90° turn in order to see the pictures (and the data) in the same orientation Many thanks !!!
maybe scipy can help https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.rotate.html ?
OK thanks ! ill try it for the image but I also will need to rotate the spatial data
hello Giovanni, do you think I need to turn the spatial transcriptomic data in order to perform batch correction with MNN on the 2 data than are not spatially aligned ? Many thanks ! Sophie
you don't need to do so, only thing is to rotate the spatial coordinates in adata.obsm["spatial"] and the image. MNN only operates on gexp counts.
did scipy.rotate work?
ah OK thanks So I rotated -adata.obsm["spatial"] -adata.uns["spatial"] ["xxx"]["image"][....] and the results is really strange I lost all the picture (it's completly zoomed on 1 corner) I'll get back to you with pictures asap thanks for your help ps maybe I dont need to rotate the image for to use MNN ???
@sophieRAIBAUD I believe you don't need to rotate the image for the MNN. I'd be curious to see whether the image and coordiantes before rotation are nonetheless aligned?
Hello Giovanni,
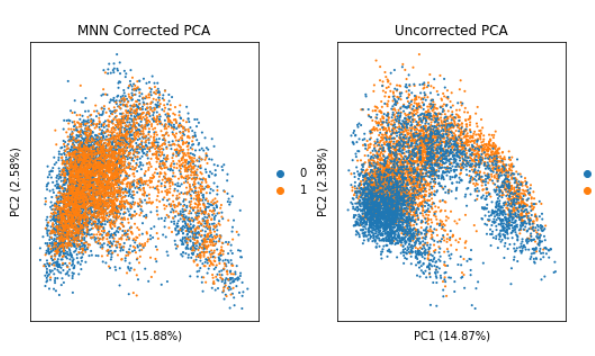
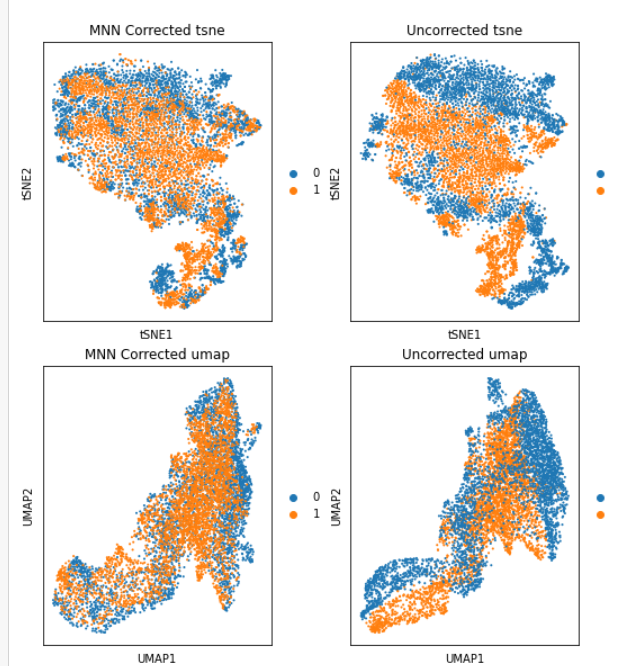
I have performed MNN on my 2 samples following this tutorial:
https://nbisweden.github.io/workshop-scRNAseq/labs/compiled/scanpy/scanpy_03_integration.html
on the PCA, tsne, and umap graphes, it look pretty cool:
but now I would like to perform a louvain clustering on corrected data
but I lost the image in the cdata:
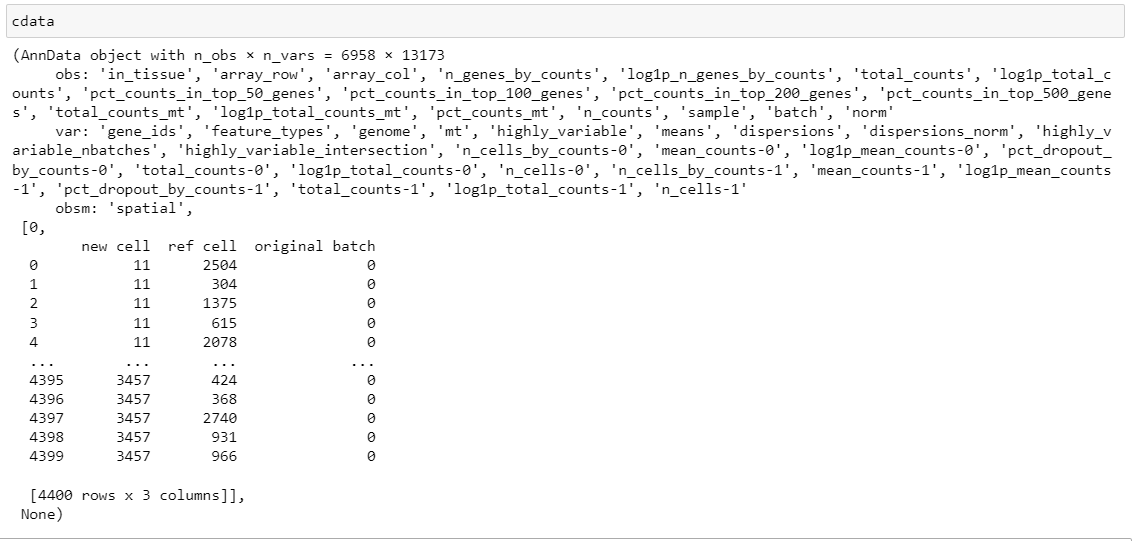
I am not sure I am doing the right thing Many thanks for your help ! Sophie
I found the solution with this notebook https://nbisweden.github.io/workshop-scRNAseq/labs/compiled/scanpy/scanpy_07_spatial.html but I had to get back the image data: corrected.uns['spatial'] = adata_raw.uns['spatial'] it works !
@sophieRAIBAUD I believe you don't need to rotate the image for the MNN. I'd be curious to see whether the image and coordiantes before rotation are nonetheless aligned?
@giovp next, I'll do it with other data that are not aligned at all thanks
Hi @sophieRAIBAUD
sorruy for late reply, I think the results looks good. could I ask you to post this type of questions in https://discourse.scverse.org/ with squidpy tag? issues are more for code bugs than best practices and usage. Thank you!