Erqiang Hu
Erqiang Hu
Please provide repeatable examples(with your data) and use the latest version of `enrichplot`. 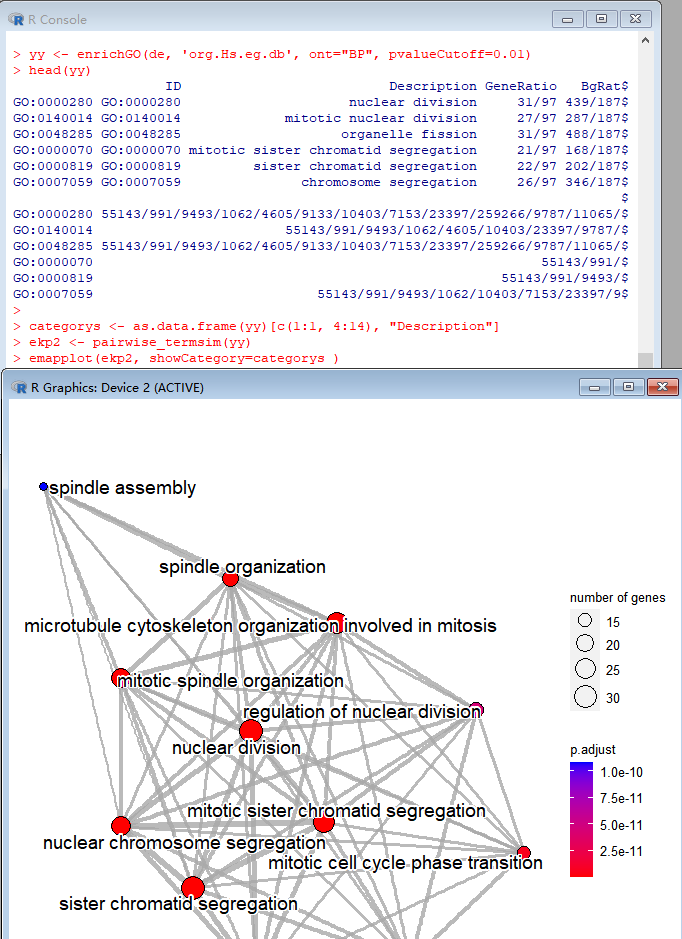
@ppdpg  ```r categorys
It is a typo in the example: should be "lung non-small cell carcinoma", not "non-small cell lung carcinoma". We will fix the document in the next version. Thanks.
It will be fixed in the next version of `enrichplot`.
For compareClusterResult object, a gene may have multiple logFCs, therefore, it is not appropriate to use them to color. So we use colors to represent the source clusters of genes.
Please provide repeatable data and code
You can use this code to make the results consistent every time: ```r # It is also correct to use other seed values set.seed(123) ```
Use the same seed before each plot, not just before the first plot: ```r library(DOSE) data(geneList) de
Similar(Semantic similarity or other calculation methods) GO terms are grouped together using K-means.
For more details, pls read the source codes.