glmpca
 glmpca copied to clipboard
glmpca copied to clipboard
Hi @willtownes, My typical Seurat workflow for multiple samples and conditions is to run PCA followed by CCA-based integration (now `IntegrateLayers` in Seurat v5), then identify one joint set of...
Hi, I found out something strange when using batch correction. These are the two result from Muraro (pancreas) and MacParland (liver) datasets. This is from Muraro 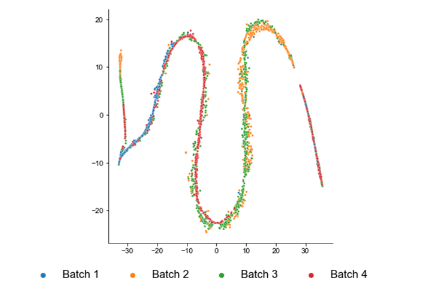 and this is...
Dear Will, I currently work with a dataset containing both gene expression data and surface marker information, using cite-seq. Now I wonder if you think there is any inherent problem...
Will, great package! I have a conceptual question about GLM-PCA with covariates: when I have sample (or gene) covariates, is GLM-PCA estimating a factor model on the variance that is...
Hi Will, Do you have any suggestions on how to choose the number of principal components in GLM-PCA? Is there a way to quantify the contributions of each PC similar...
Since the original publication is called _Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model_, it would be nice to have a part of the vignette...
In the [Collins paper](https://papers.nips.cc/paper/2001/file/f410588e48dc83f2822a880a68f78923-Paper.pdf) they write Would it be possible to add the ability for each column in Y to be distributed by a different exponential family distribution?
As pointed out by https://github.com/willtownes/scrna2019/issues/5 glmpca is returning factors as a data frame instead of a matrix (as the documentation claims), which causes downstream problems converting SingleCellExperiment objects to Seurat....
The glmpca_family object consumes a lot of memory because it preserves the environment where `infograd` and `dev_func` functions are defined. Refactor this code so these functions depend only on their...
It would be great to use a trained glmpca to infer factors from new data. Possibly like: `new_factors
