saras224
saras224
Hi @donovan-parks! I just ran into an error while running the gtdbtk classify_wf command. My command was: **gtdbtk classify_wf --genome_dir /lustre/rsharma/FLOOR-METAGENOME-NOVASEQ/ASSEMBLY/MAXBIN_REZ/GOOD_QUALITY_BINS/GBINS/ --out_dir /lustre/rsharma/FLOOR-METAGENOME-NOVASEQ/GTDBTK_REZ/** Terminal output: 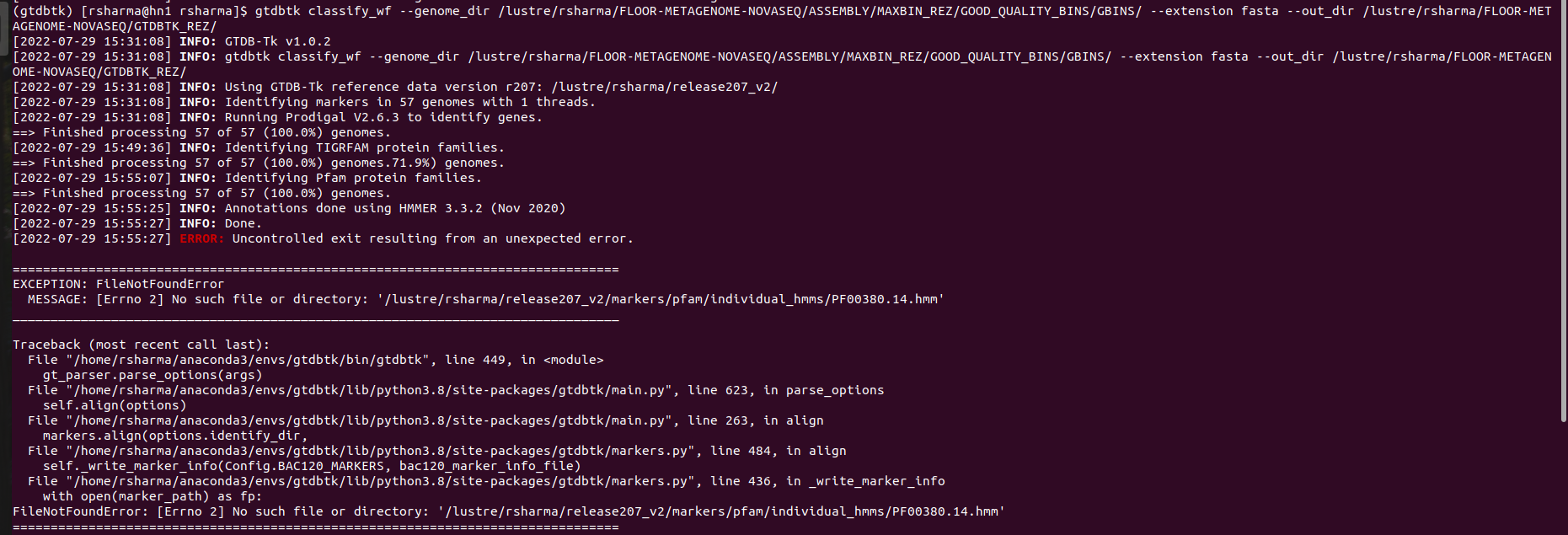 ERROR:...
Hey @allind I tried to install eukdetect but when I run this command: conda env update --name Eukdetect -f environment.yml it gave an error saying: SpecNotFound: Invalid name, try the...
Hi @wwood I just want to know how can I change the lables of the x axis in the results of the roary plots? I want to label it according...
Hi @rmFlynn ! After the processing of the databases can I delete these database files listed below? 16GB Pfam-A.full.gz 2GB kofam_profiles.tar.gz 42GB uniref90.fasta.gz 1GB viral.merged.protein.faa.gz 1GB kofam_ko_list.tsv.gz 1GB merops_peptidases_nr.faa 1GB...
Hi @rmFlynn @ileleiwi @rdenise I ran DRAM for 9 MAGs and it gave me an **error that the object has no attribute query**, can you please go through the log...
Hi @mrobbert While running the DRAM annotation command I encountered this error:  Please help me resolve the error so that I can proceed further. Thanks in...
Hi! @rmFlynn I have downloaded database files but I do not know how to provide database location to the database_handler.py script. When I run the analysis I get the following...
[GTDB_TAXONOMY.xlsx](https://github.com/fbreitwieser/pavian/files/10684874/GTDB_TAXONOMY.xlsx) Hi @fbreitwieser I wanted to ask if pavian will be able to generate a Sankey diagram for the GTDBtk taxonomy file. I am providing the gtdbtk report file in...
Hello @biofuture just before creating the final output result it gave this error: Error in tempdir() using /tmp/XXXXXXXXXX: Could not create directory /tmp/EI0zRIUpTg: No such file or directory at /usr/local/share/perl/5.26.1/Parallel/ForkManager.pm...
Hi @biofuture I have used ARGs-OAP successfully to obtain the ARGs profile for all of my metagenomic datasets and now I wanna process them further and do the following: -...
