Panu Lahtinen
Panu Lahtinen
Hi, I worked on Pyresample gradient search resampler (originally implemented by @mraspaud) in https://github.com/pytroll/pyresample/pull/282 a while back. The resampler now maps the source chunks to those, and only those, target...
I thought we already did select the closest TLE in time, but apparently not: https://github.com/pytroll/pyorbital/blob/master/pyorbital/tlefile.py#L197-L201
I guess the flat 100 % in the beginning is the Satpy file handler creation we've already discussed in https://github.com/pytroll/satpy/issues/2186 ?
Thanks for testing! I noticed the same increase in processing time for local files, but didn't yet have time to look into it. My guess is some `da.array()` calls or...
See if https://github.com/pytroll/satpy/pull/2096 provided something that is more usable for you? With it, this should work: ```python filenames = [ 'simplecache::s3://satellite-data-eumetcast-seviri-rss/H-000-MSG3*-202204260855-*', ] scn = Scene(reader='seviri_l1b_hrit', filenames=filenames) scn.load(list_of_datasets) ``` Some additional...
What if you don't use `glob` directly, but just pass the glob pattern as filename like I showed above?
Same here. Just did `mamba update --all` and updated to latest Satpy `main` and the whole example script (the `I01` variant) takes only 3 seconds to run.
The same is seen when loading a bunch (20) composites instead of the single one.
Instead of resampling and saving, doing ```python glbl['ir_105'].compute() ``` shows the same performance issue: 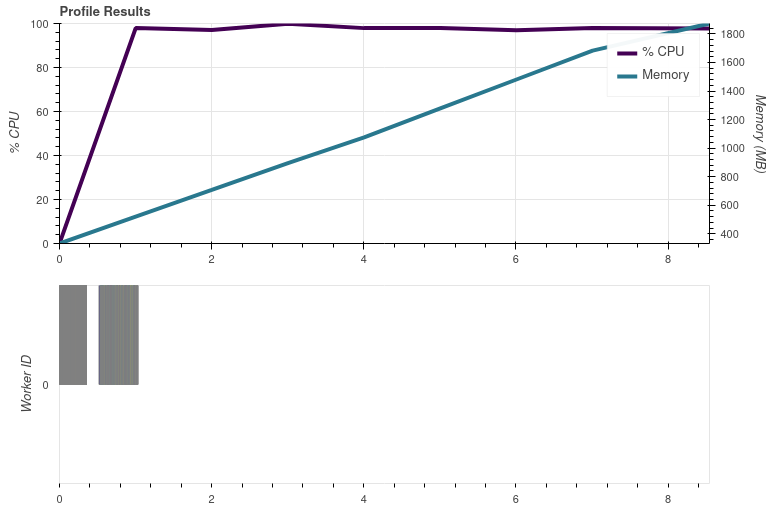
ABI processing is done in parallel, so at least it isn't the NetCDF C library that is stopping the parallel processing. And the FCI data are processed in single thread...