Merlin Szymanski
Merlin Szymanski
Ah nice, so its only a matter of time then :) Thanks for looking into it!
Is already part of the dsl2-metagenomics branch https://github.com/nf-core/eager/pull/1019
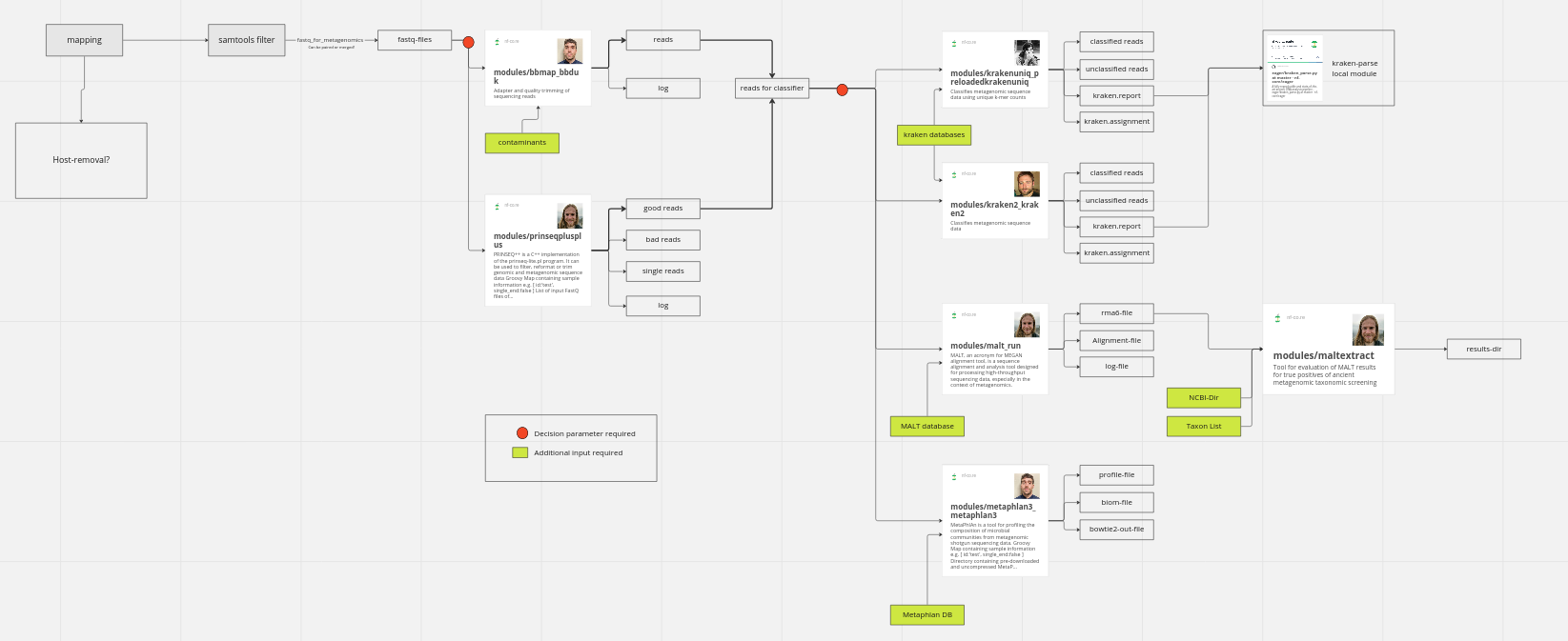 Hey, I outlined the metagenomics-pipeline. If you have any comments/ things to add, please let me know!
Current progress: 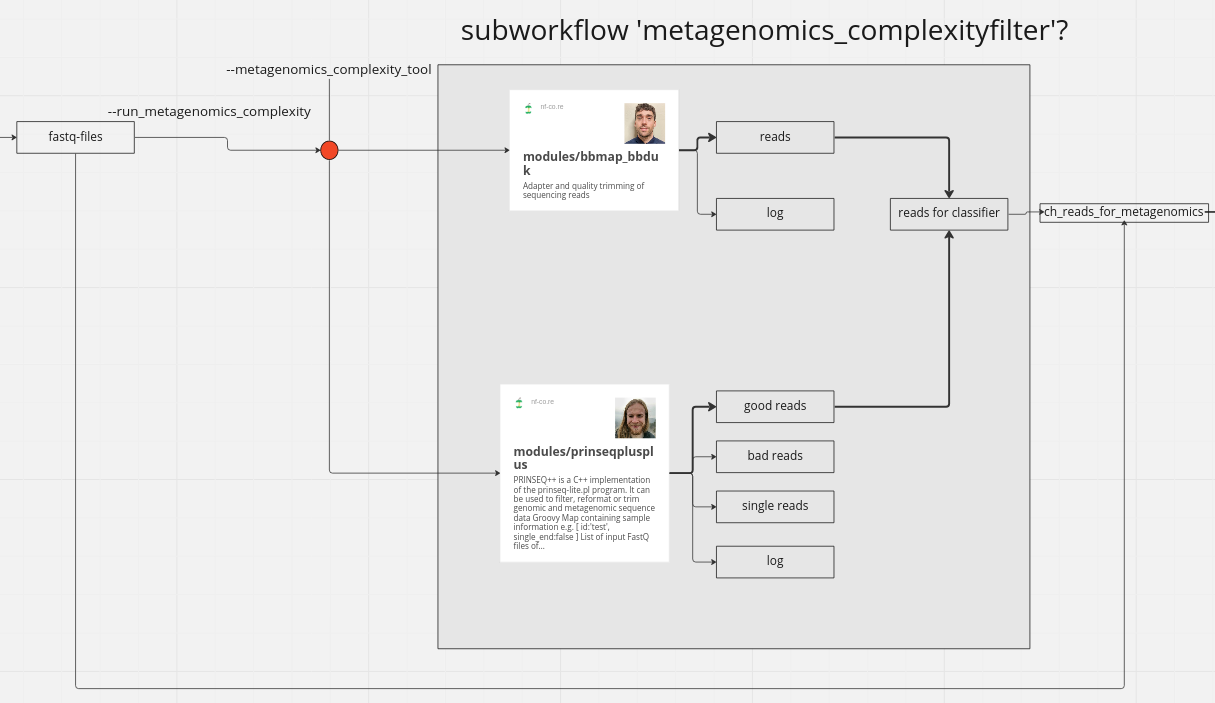
@ilight1542 The current version in dsl2-metagenomics works for the following tests [tests.md](https://github.com/nf-core/eager/files/11847236/tests.md)
closed with #1098
@ilight1542 maltextract+AMPS works now, however, there are many optional parameters, so I'll do the comprehensive testing on friday.
All tests, except Metaphal have passed today (see file attached). [tests.md](https://github.com/nf-core/eager/files/12829851/tests.md) ToDo for the next testing: [] optional parameters [] check the expected output [] update the manual_tests.md file I'm...
> RE: keeping strandedness. Since the only meta in malt-run is the meta with the list of read files, keeping info on which samples have single-stranded library prep must be...
> > RE: keeping strandedness. Since the only meta in malt-run is the meta with the list of read files, keeping info on which samples have single-stranded library prep must...