Chris Gorgolewski
Chris Gorgolewski
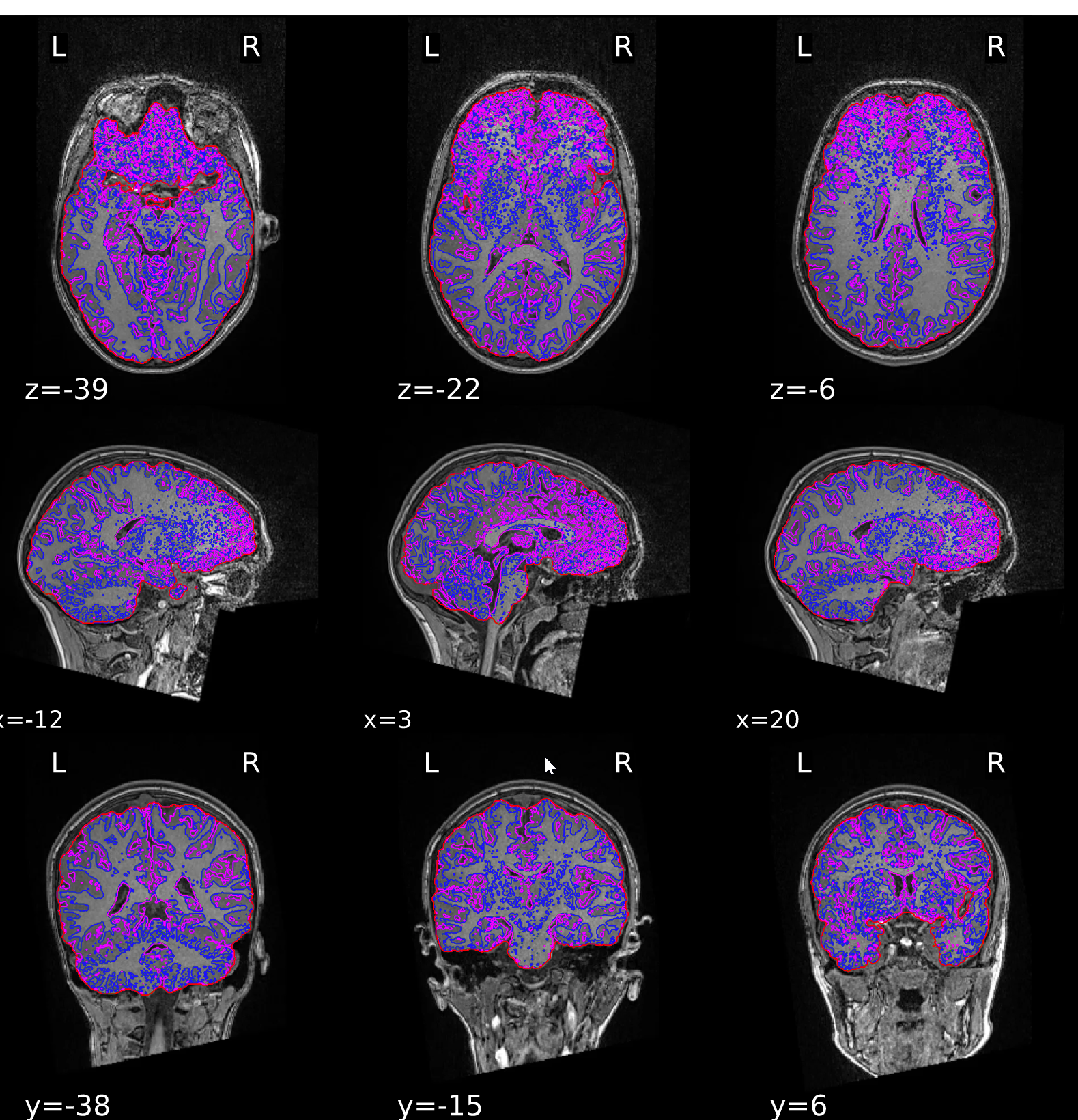 ds000200/reportlets/fmriprep/sub-2014/anat/sub-2014_T1w_seg_brainmask.svg
Last case had a negative influence on surface reconstruction 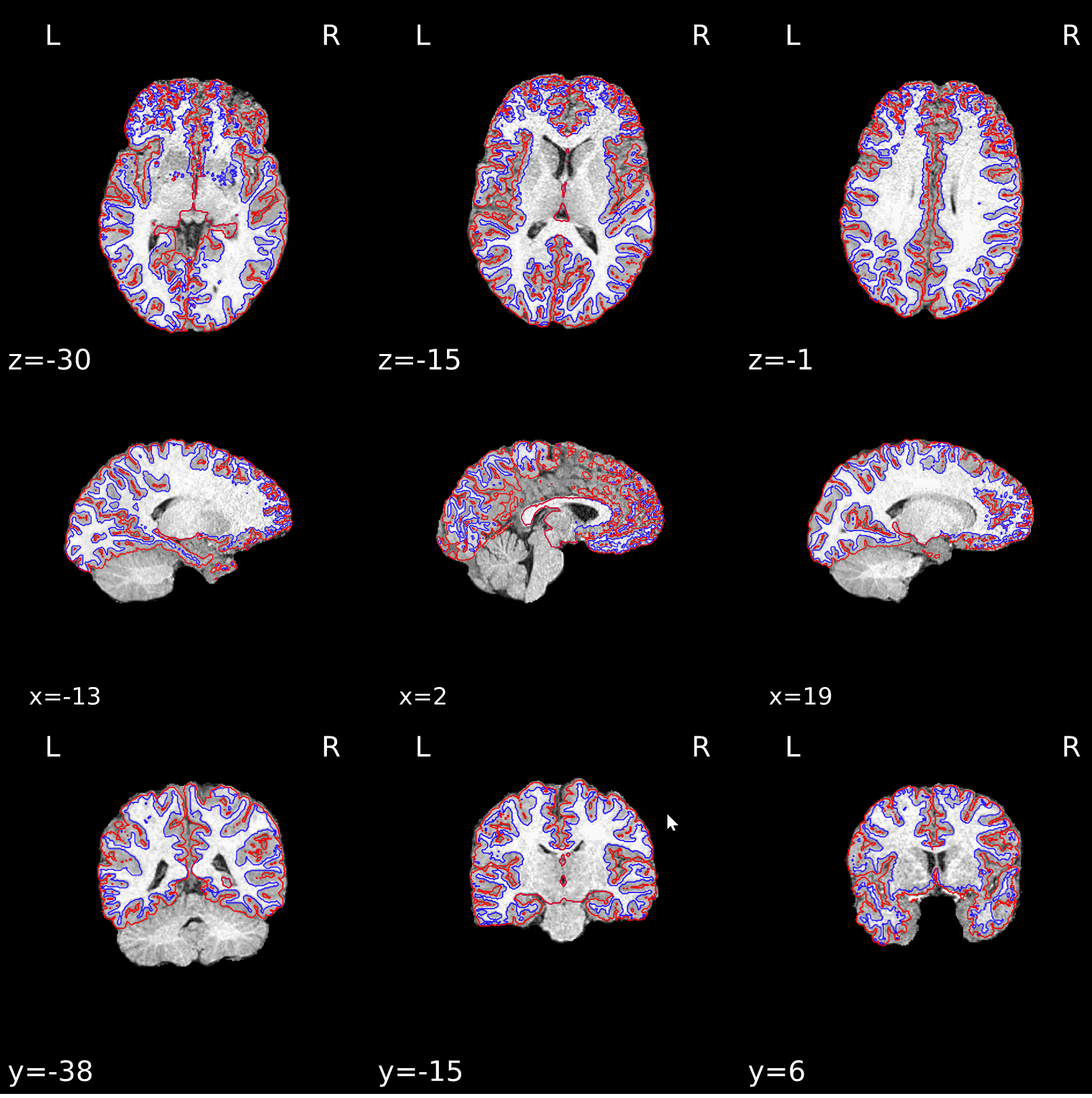
Python implementation (with some potential speed improvements): http://scikit-image.org/docs/dev/auto_examples/filters/plot_nonlocal_means.html
DenoiseImage results are pretty impressive - sadly it took over 2h to run 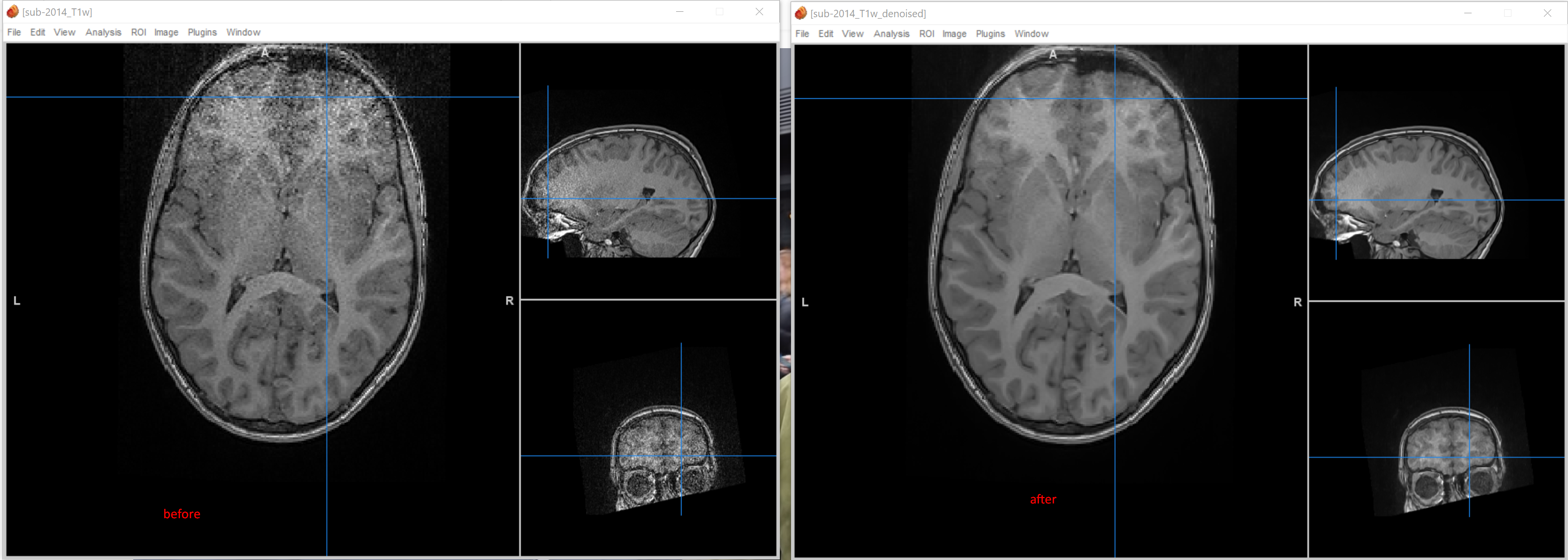
Another option is using a skullstripping procedure designed for MP2RAGE such as the one implemented in nihighres ``` skullstrip_results = nighres.brain.mp2rage_skullstripping( second_inversion=dataset["inv2"], t1_weighted=dataset["t1w"], t1_map=dataset["t1map"], save_data=True, output_dir=out_dir, file_name="sub001_sess1") ``` More info...
A pull request would be awesome (I am not currently working on this). Assuming the data will be formatted according to [BEP001](https://docs.google.com/document/d/1QwfHyBzOyFWOLO4u_kkojLpUhW0-4_M7Ubafu9Gf4Gg/edit#heading=h.7m77cpb9r0n9) is the way to go. Thank you in...
+1! On Sun, Jan 13, 2019, 7:13 PM Chris Markiewicz Lesion masks are actually derivatives, so instead of looking for > _roi.nii.gz files, we should be accepting a derivative root...
@AllenChong "PowerKill Project"? What do you mean?
Awesome @eayoungs - looking forward!
I prefer an asynchronous and open mode of communicating via GitHub comments/issues, but I will keep checking the slack as well.