Chris Friedline
Chris Friedline
Yeah, same as my #29 and @nileshpatra's #139. There are many use cases for being able to cleanly diff code between tags and know what's changing, especially with production use....
Here is some code to reproduce: `test_df.tsv`: https://drive.google.com/file/d/1kmaJGb7UP1UO8BtsVgBeCFDDyIXLs9SF/view?usp=sharing `test_exons.bed`: https://drive.google.com/file/d/1tipD2PVo66eU96Y2m-g82fzUoT0ZwCkB/view?usp=sharing ``` import pandas as pd import pyranges as pr test = pd.read_csv("test_df.tsv", sep="\t") test_exons = pr.read_bed("test_exons.bed") test_df = test[ (test.vendor...
@endrebak In case you can't replicate it, here's another experiment. Using data from `test2`, and overlapping the same ranges, it only works in the second case if I recreate the...
I'm working on recreating this now, though looking at the changes, I'm not sure it will make much of a difference.
I'm sad to report that the environment I was testing in, which also was a production analysis environment, is no longer exhibiting this behavior. There have been so many changes...
Wait, I may have recreated it just now. ;-)
Yup - as expected, even with 0.0.88, without chromosome as a category type in the df for `pr.PyRanges()`, the previously failing joins still fail.
Some more info here where my case is failing - I'm working on an example to provide. This is with the latest 0.0.88. I'm grouping a data frame as follows...
Looking more closely at my `failed_loci_per_assay_df` data frame, the `Chromosomes` type is not just `object`, but `mixed`. This is likely my issue, and why it can't be replicated by simply...
Stepping through the code, it is indeed creating category, but not of `object`, but instead it's `int64` For example, in `init.py` 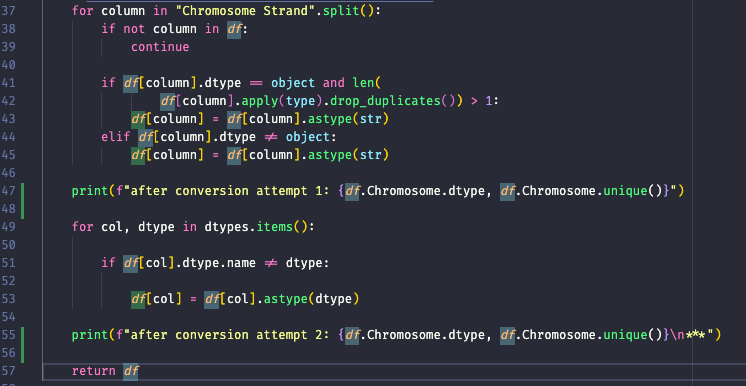 For a working join, I get this: 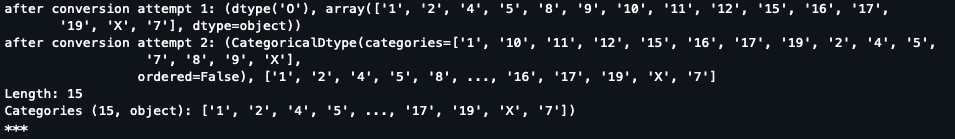...