Jiangjiangzhang6
Jiangjiangzhang6
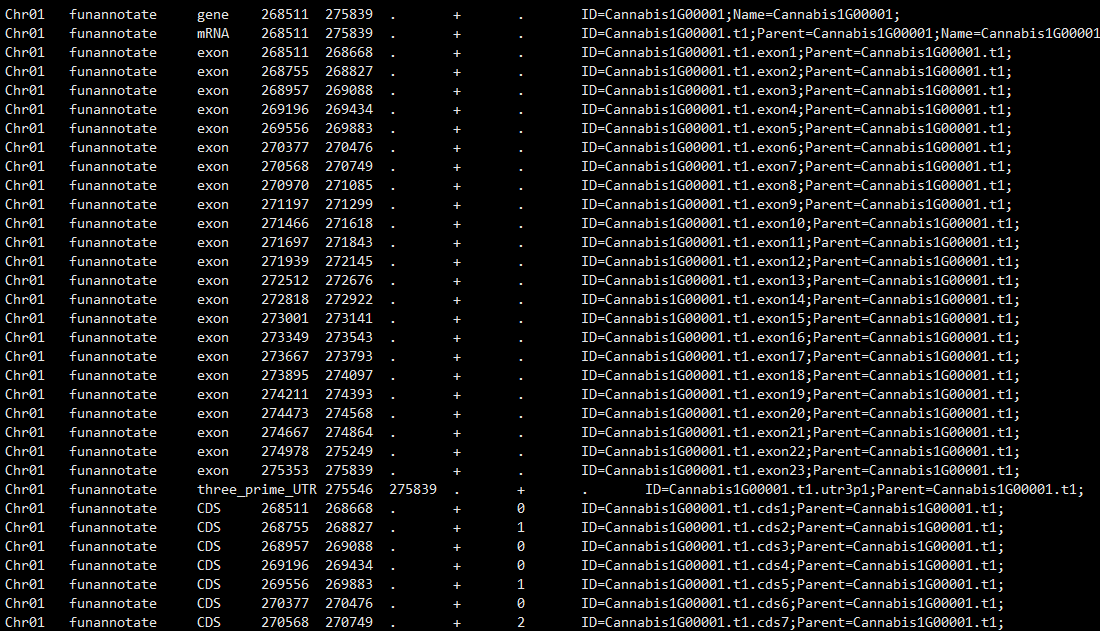
> Hi @Jiangjiangzhang6 , > > When parsing your gff emapper is expecting a tab delimited file with 9 fields, as in https://www.ensembl.org/info/website/upload/gff3.html > > Is it your gff file...
its show 0 and 9 
maybe I know error , its would not allow blank line
thank you for your reply , now submit it to the cluster, thank you
 wheather the genome is to big so our software could not handle with it
 and the assembly file could not load in , any other way to solve this
> > the genome was ultra large scale about 23G, after I motif the assembly via the juicerbox, and conduct the next step with the command "bash ~/software/3d-dna-master/run-asm-pipeline-post-review.sh -r ../fusion_second2.0.review.assembly...
conda install -c conda-forge gxx did it the error casued by the g++ complier
version: 180922 -r|--rounds flag was triggered, will run 0 round(s) of misjoin correction. ############### Starting iterating scaffolding with editing: ...starting round 0 of scaffolding: :) -p flag was triggered. Running...