MolecularNodes
 MolecularNodes copied to clipboard
MolecularNodes copied to clipboard
Toolbox for molecular animations in Blender, powered by Geometry Nodes.
Some node groups are not pre-defined, to allow for custom inputs such as a chain selection, chain color and other structure-specific nodes. This allows one chain selection node to have...
Hi Brady, I would like to add a new Material called "MN AO Emission". 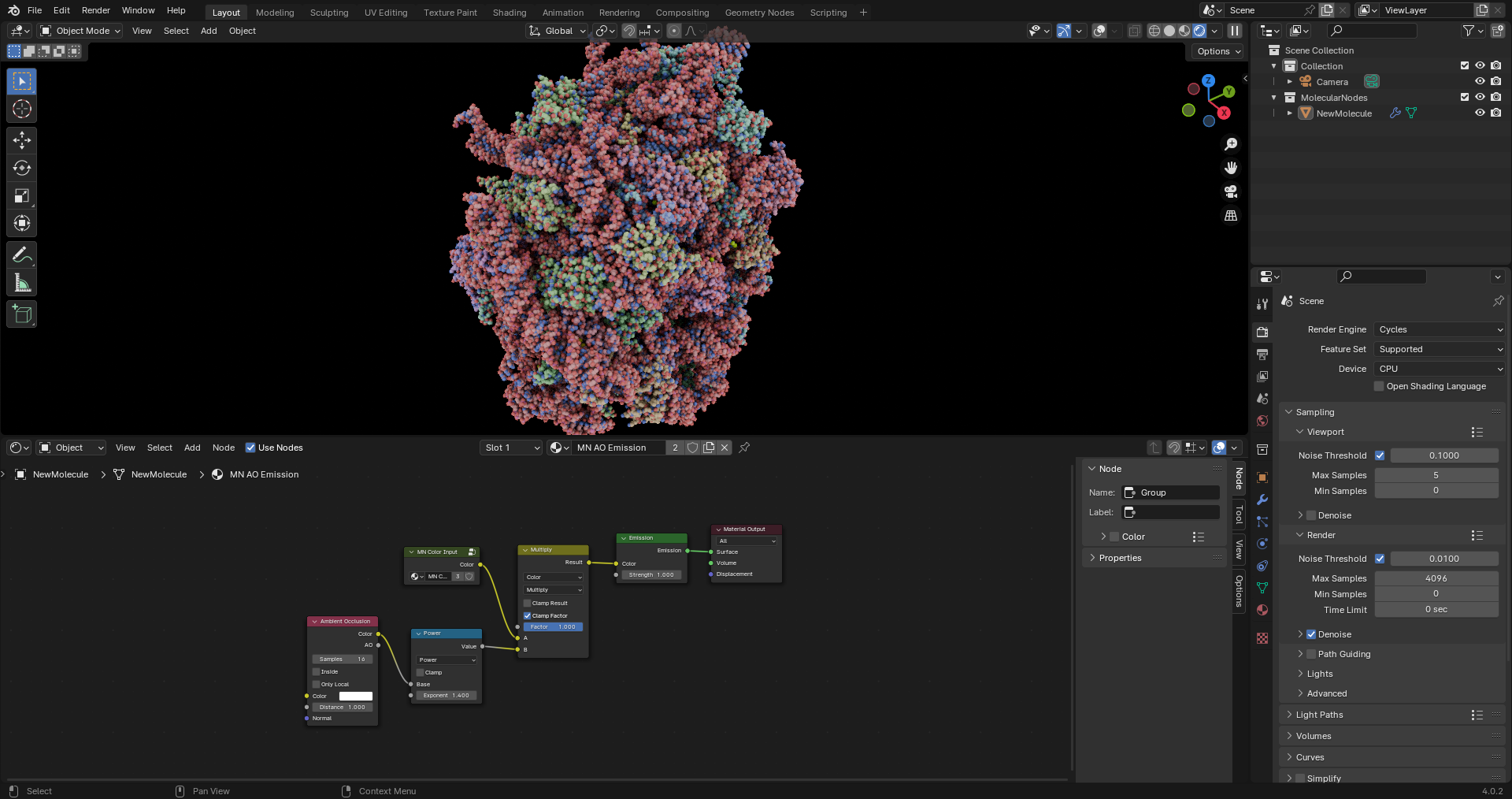 I use it a lot in my work, mostly because it makes renders work without...
When importing a structure either locally or from the PDB, a potential option to remove hydrogens as most protein structures don't want to include hydrogens when visualising the structures.
This should add the starfile node to the documentation. I am still working on the tutorial, but figured I open the PR to see if the CI for the docs...
For instancing of particles, reads `.ndjson` files and creates points for instancing. Currently just the initial barebones importer which can parse the positions and the rotations for the points. Doesn't...
Proposal for Issue #464 Changes: - abc for molecules - molecule classes that want to inherit from it require two methods: ```python @classmethod @abstractmethod def _get_structure(cls, fpath: str): ... @abstractmethod...
Related to #465, this adds a button in the add-on preferences that the user can click to register the add-on's assets with Blender's asset library browser.  Allows for quicker...
In the very early stages, but there is currently a new molecular dynamics database being built, modeled on the PDB. This will allow downloading and streaming of MD trajectories via...
**Describe the bug** topology and trajectory created with recent version of oxDNA 3.6.1 does not load in MN **To Reproduce** [http://tacoxdna.sissa.it/XYZ_oxDNA] - generate test.oxdna using tacoxdna [https://lorenzo-rovigatti.github.io/oxDNA/relaxation.html] - First stage:...
As described in the issues #454 and #459, the molecule class can be optimized and unified with the AtomGroupInBlender class. Goals: - [x] clean up s. #454 - [x] write...
