Obs image layer is flipped in Napari
... Hi all,
I was trying to use interactive mode with Napari. everything worked fine except when I try to plot genes or observations on top of my segmentation image, spots look to be flipped on the Y axis but couldn't find where it gets flipped.
Using Squidpy from github main and Napari 0.4.17
Hi @Tonyspuri , can you please provide a minimal reproducible example?
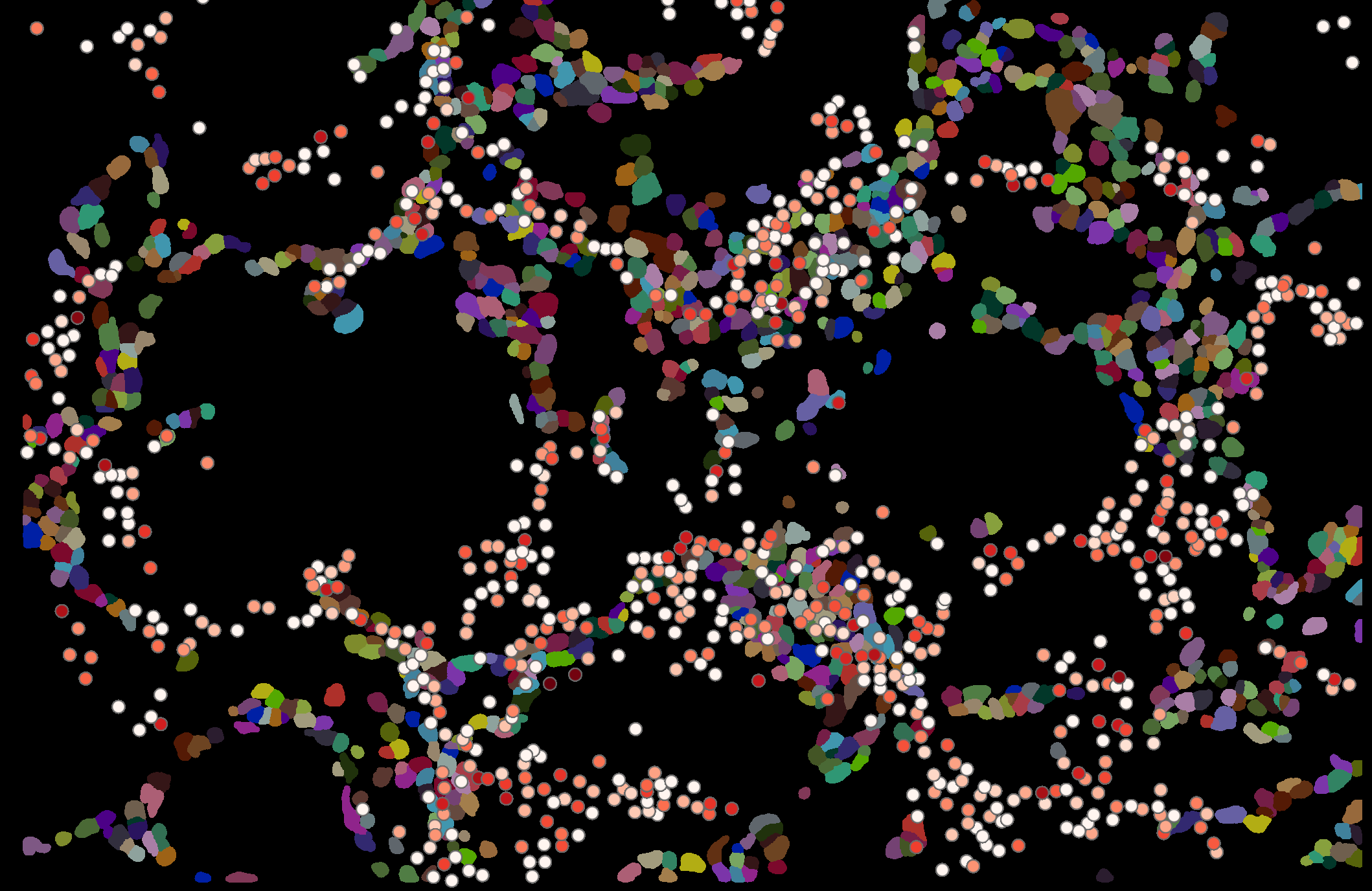
I was playing with Nanostring Spatial data mostly following the tutorial to read in the files, quality control etc.
Then I tried to run interactive mode, first I was setting up imagecontainer
imgs = []
for library_id in adata.uns["spatial"].keys():
img = sq.im.ImageContainer(adata.uns["spatial"][library_id]["images"]["hires"], library_id=library_id)
img.add_img(adata.uns["spatial"][library_id]["images"]["segmentation"], library_id=library_id, layer="segmentation")
img["segmentation"].attrs["segmentation"] = True
imgs.append(img)
img = sq.im.ImageContainer.concat(imgs)
and run interactive mode
viewer = img.interactive(adata, library_key='fov', library_id="1")
Hires image and segmentation image overlay perfectly well but when I try to add Obs or genes layer it basically flips on the y axis.
hi @Tonyspuri ,
this is because the obsm["spatial"] coordinates are flipped on the Y axis I believe. Think the best way is to flip them yourself manually. We won't support this napari implementation anymore as in few weeks we'll have a new format and napari plugin for scverse. Will try to remember to report here relevant documentation.
@giovp Great, thanks a lot! I'll keep an eye out for a new format but shortterm try to flip the coordinates
you can check the new spatialdate ecosystem for nanostring here: https://spatialdata.scverse.org/en/latest/tutorials/notebooks/notebooks/examples/technology_cosmx.html