how to apply custom transformation to image crops before feature extraction
Hi, thanks for the awesome tool. I am interested in doing some kind of normalization/custom transformation on the image before I run feature extraction (calculate_image_features). But all of my attempts so far have resulted in out of memory during run time.
For example, suppose I want to do a minmax scaling on the image. My attempt included:
import dask as da
import dask_image.imread as imread
import squidpy as sq
dimg = imread.imread(img_path)
x_min = da.array.min(dimg, axis=(0,1,2))
x_max = da.array.max(dimg, axis=(0,1,2))
dimg = (dimg - x_min) / (x_max - x_min)
img = sq.im.ImageContainer()
img.add_img(dimg, layer="image")
and then using calculate_image_features with img.compute(), but this results in out of memory.
I have also tried
img.apply(lambda x: da.array.true_divide(da.array.subtract(x, min_val),
da.array.subtract(max_val, min_val)), lazy=False, copy=False, layer='minmax')
to just create a new layer from the original img, but this runs into out of memory also (immediately if I set lazy=False and during feature extraction if I set lazy=True). Any suggestions how to accomplish applying a custom transformation to each image crop either during or before feature extraction? Thanks a lot!
Hi @fedshyvana ,
I'd try tweaking the parameters of a local dask cluster, e.g.:
from dask.distributed import Client
# register client
client = Client(n_workers=4, threads_per_worker=2, memory_limit='8GB', memory_target_fraction=0.95)
print(client.dashboard_link) # dashboard location to monitor workers
This depends on how large your image is, how large your image chunks are and how much memory you have available (the 'memory_limit=...' worker-specific).
In my case, I didn't get any memory issues (the image was just gaussian noise of shape (100_000, 100_000, 3), chunks were of size (10_000, 10_000, 3) [in total 100 chunks] and I have 32GB RAM).
thank you! ill give it a try today.
@michalk8 hi, sorry I am not that familiar with dask, but after using dask_image.imread to read in my image I see that it only has one chunk:
| Array | Chunk |
|---|---|
| 2.54 GiB | 2.54 GiB |
| (1, 28672, 31744, 3) | (1, 28672, 31744, 3) |
| 3 Tasks | 1 Chunks |
| uint8 | numpy.ndarray |
So to be able to use dask.distributed for parallel compute I would need to first chunk the image? Or does Squidpy automatically chunk the image depending on the analysis required (e.g. when calling calculate_image_features with a specified patch size)?
On a related note I see that the image wrapped inside a Squidpy ImageContainer object layer is an instance of xarray.DataArray, is that just another wrapper around dask.array?
Hi @fedshyvana ,
you can initialize the ImageContainer also from a file as:
import numpy as np
import squidpy as sq
from PIL import Image
Image.fromarray(np.random.normal(size=(1000, 1000))).convert("RGB").save("test.jpeg")
img = sq.im.ImageContainer("test.jpeg", lazy=True, chunks=(1, 500, 500, 3), layer="test")
img["test"] # verify that it's correctly chunked
Please, also take a look at the documentation of the ImageContainer here (or here).
Or does Squidpy automatically chunk the image depending on the analysis required (e.g. when calling calculate_image_features with a specified patch size)?
No, we don't automatically select chunk sizes anywhere, user has to select them based on their use-case and environment, see also this short Twitter thread (afaik a blog post with more details will come soon).
On a related note I see that the image wrapped inside a Squidpy ImageContainer object layer is an instance of xarray.DataArray, is that just another wrapper around dask.array?
Yes, it's just a wrapper that can use numpy.array/dask.array/etc as the backend.
Hi @michalk8, thanks a lot for the response! I have revised my workflow based on your suggestion:
# initialize cluster
cluster = LocalCluster(
n_workers=4,
threads_per_worker=2,
memory_limit='8GB'
)
client = Client(cluster)
print(client.dashboard_link)
# some example values for min-max scaling
min_val = np.array([0,0,0])
max_val = np.array([255, 200, 255])
scale = max_val - min_val
# load an image into ImageContainer, specifying chunksize
img = sq.im.ImageContainer(img_path, lazy=True, chunks=(1, 512, 512, 3), layer="image")
# queue-up minmax scaling transformations
img.apply(lambda x: x - min_val, layer='image', lazy=True, copy=False)
img.apply(lambda x: x / scale, layer='image', lazy=True, copy=False)
# compute features
sq.im.calculate_image_features(
anndata,
img.compute(),
layer=layer,
features="summary",
key_added='features',
n_jobs=4,
scale=1.0,
)
Unfortunately I always end-up with issues with workers running out of memory (says worker restarting) and then a bunch of errors that eventually cause the runtime to crash.
I notice that I never reach the progress bars that sq.im.calculate_image_features normally displays for how many spots it has processed so my understanding is that the runtime never made it past the min-max transformation.
To me, intuitively this shouldn't be that difficult of a task right? We can imagine that a naive solution would be to just loop through all pixels in the image one by one, subtract the mean and scale it - so I am not sure what I am during wrong for dask to struggle so much.
Alternatively, given that I really just want to apply the transformation to the crops that correspond to the visum spots individually (which under the hood is generated by generate_spot_crops), is there a way I can let squidpy know to just do a function call (e.g. the minmax scaling) on each of the spot crop before features_summary is computed on it?
Thanks so much once again! Max
@fedshyvana could you try removing the img.compute() call? This will trigger the dask computation for all layers and materialize the arrays, which is the reason I believe it crashes/no progress bar is shown (it's mostly a convenience function for small containers).
Alternatively, given that I really just want to apply the transformation to the crops that correspond to the visum spots individually (which under the hood is generated by generate_spot_crops), is there a way I can let squidpy know to just do a function call (e.g. the minmax scaling) on each of the spot crop before features_summary is computed on it?
From the high-level sq.im.calculate_image_features, there's a possible workaround by supplying a custom function to compute your features as e.g.:
sq.im.calculate_image_features(
adata,
img,
layer="image",
features="custom",
features_kwargs={"custom": {"func": lambda arr: np.mean(arr)}},
)
See here as what function signature is expected.
However, I see the benefit of adding optional transformation before feature calculation, since this solution is applicable to only custom features - could be specified as apply_kwargs parameter. Also pinging @giovp for any thoughts.
Alternatively, you can use the container itself to generate the spot crops as, though it's a bit lower level (+ you lose the parallelization over spots, which in this case is fine - I wouldn't use it [n_jobs=1 in your above code] until the memory issue is resolved):
observations, feats = [], []
for spot, obs in img.generate_spot_crops(adata, return_obs=True):
# spot is again `ImageContainer`, obs is from `adata.obs_names`
features = ...
observations.append(obs)
feats.append(features)
adata.obsm["my_features"] = pd.DataFrame(feats, index=observations)
@michalk8 thanks! this is very helpful information! If I understand correctly, calling "compute()" on the ImageContainer loads the underlying image layers into persistent memory? Indeed I am able to run the feature extraction without img.compute(), however the subsequent feature extraction is like an order of magnitude slower though....
Is there any reason why when reading "larger than memory" images/arrays, ImageContainer/dask is so much slower than using libraries such as openslide?
I did a quick benchmark on the particular 28672 x 31744 image, reading the same 256 x 256 patch at position (0,0) 10 times into numpy arrays.
First using ImageContainer crop_corner (without .compute()).
%time
dimg = dask_image.imread.imread(img_path)
img = sq.im.ImageContainer()
img.add_img(dimg, layer="image")
for i in range(10):
patch = img.crop_corner(0,0, 256)['image'].values
if i == 0:
print(patch.shape)
Result: (256, 256, 1, 3) CPU times: user 1min 28s, sys: 1min 23s, total: 2min 51s Wall time: 2min 3s
With chunking, knowing apriori that each patch is 256 x 256:
%time
img = sq.im.ImageContainer(img_path, chunks=(1, 256, 256, 3))
for i in range(10):
patch = img.crop_corner(0,0, 256)['image'].values
if i == 0:
print(patch.shape)
Result: (256, 256, 1, 3) CPU times: user 28.5 s, sys: 26.8 s, total: 55.4 s Wall time: 39.3 s
Next by first calling .compute()
%time
img = sq.im.ImageContainer(img_path, chunks=(1, 256, 256, 3))
img.compute()
for i in range(10):
patch = img.crop_corner(0,0, 256)['image'].values
if i == 0:
print(patch.shape)
Result: (256, 256, 1, 3) CPU times: user 9.98 s, sys: 5.31 s, total: 15.3 s Wall time: 10.7 s
Indeed this is much faster but requires much more memory.
Lastly with openslide, which does not load the whole image into memory:
%time
import openslide
wsi = openslide.open_slide(img_path)
for i in range(10):
patch = np.array(wsi.read_region((0,0), 0, (256, 256)))
if i == 0:
print(patch.shape)
Result: (256, 256, 3) CPU times: user 20.4 ms, sys: 4.7 ms, total: 25.1 ms Wall time: 24.3 ms
As you can see openslide is very fast despite not needing to load the whole image into memory. Is there any way we can achieve similar performance with dask/ImageContainer?
Thanks again for all the detailed responses - greatly appreciated!
dimg = dask_image.imread.imread(img_path)
Small heads-up: problem with dask_image.imread.imread is that at least for JPEGs, it loads the data into memory just to get the shape of the image (that's why we don't use it to read the images, just daskify skimage.io.imread).
As you can see openslide is very fast despite not needing to load the whole image into memory. Is there any way we can achieve similar performance with dask/ImageContainer?
Pretty impressive speed-up, will have to check-out openslide, thanks a lot for pointing it out!
There's 1 issue with:
img = sq.im.ImageContainer(img_path, chunks=(1, 256, 256, 3))
for i in range(10):
patch = img.crop_corner(0,0, 256)['image'].values
if i == 0:
print(patch.shape)
is that acessing .values will trigger the computation in the dask graph, whereas we could first define the graph and exploit the parallelism as e.g.:
img = sq.im.ImageContainer(img_path, chunks=(1, 256, 256, 3))
arrs = []
for i in range(10):
arrs.append(img.crop_corner(0,0, 256)['image'].data) # using .data instead of .value
da.stack(arrs).mean(0) # just an example
On some toy data, this has tripled the performance:
from 285 ms ± 16.7 ms per loop (mean ± std. dev. of 7 runs, 1 loop each) to 108 ms ± 3.08 ms per loop (mean ± std. dev. of 7 runs, 10 loops each) (using random 32k x 32k array from dask).
In squidpy.im.calculate_image_features, we do it however as in the 1st case and I should look into whether we can benefit from speed-ups there if the underlying array is from dask (for now, the improved behavior can be implemented using img.crop_spot_generator).
Also, note that chunks in squidpy.im.ImageContainer during loading just means re-chunking (for subsequent convenience). Afaik dask-image also can't load x/y chunks from file (though will parallelize if you have more slices in a TIFF); I am also not aware (apart from your approach with openslide) of any dask-related approach that would be able to do so (would be great though).
That being said (and as well as from the result on the random array), the bottleneck seems to be the read + possible lots of rechunking (using chunks='auto', I get the below graph as img["image"].data[:256, :256].visualize().
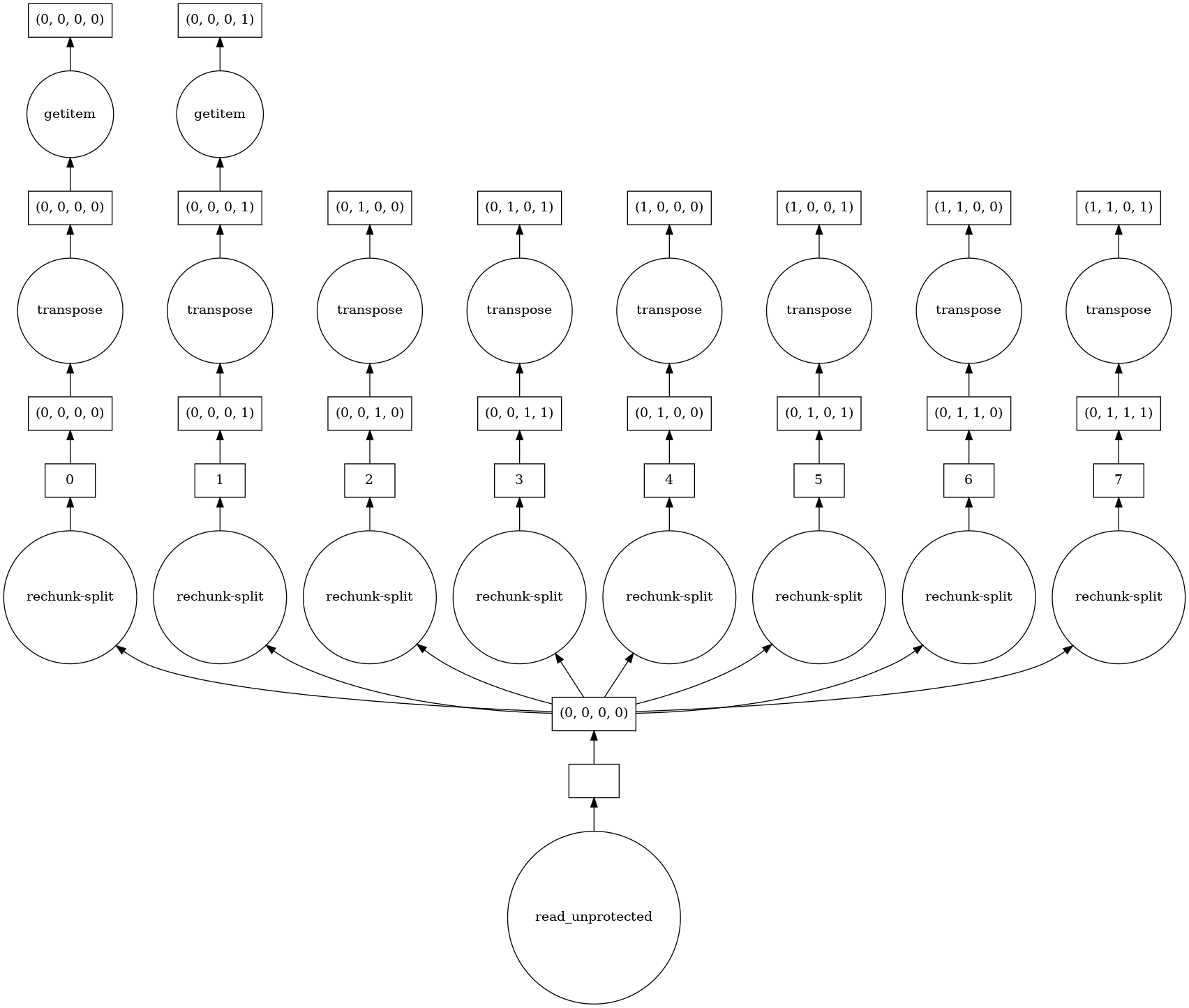 To further quantify the
To further quantify the chunks effect, I ran it with:
chunks=None: 12.8 s ± 417 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
chunks='auto': 14.6 s ± 653 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
chunks=(1, 256, 256, 3): 17.4 s ± 877 ms per loop (mean ± std. dev. of 7 runs, 1 loop each)
In summary, IO can be improved, smaller chunks in the read in the above examples make it worse and defining computational graph where individual work can be done in parallel by dask helps the performance. Depending on how easy it would be to integrate openslide, we could include it (at the cost of losing the comp. graph + parallel execution from dask, unless we can wrap it somehow).
@michalk8 thanks so much for the explanation! So for starters I don't come from a software engineering background so I am generally not capable of identifying the most efficient implementation. But for reference, openslide is something that the computational pathology community has used extensively for reading multi-resolution, larger than memory, gigapixel images.
What I have found to work for my projects in the past, is that if I know a priori how many locations I want to read (e.g. I know I have 2000 spots in total that i need fetch from the image), I just instantiate a single openslide object, and wrap it in a map-style dataset that maps indices to spot coordinates (i.e. the index 0 corresponds to coordinates for 1st spot, etc.) for example https://pytorch.org/docs/stable/data.html#map-style-datasets. And then use a multi-threaded dataloader where now each worker can just go in, given an index, call on the read_region function of the openslide to fetch a patch at the corresponding coordinate. e.g. https://pytorch.org/docs/stable/data.html#torch.utils.data.DataLoader. With many workers I was able to parallelize the operation of reading a batch of patches from a single larger than memory image. And in the dataset, I could also define some custom transformation I want to apply to each patch independently before it's returned.
Example code might look like:
import torch
import openslide
import numpy as np
class img_dataset(torch.utils.data.Dataset):
def __init__(self, coords, img_path, patch_size=256, custom_transform=None):
self.coords = coords
self.wsi = openslide.open_slide(img_path)
self.f = custom_transform
def __len__(self):
return len(self.coords)
def __getitem__(self, index):
patch = np.array(self.wsi.read_region(tuple(self.coords[index]), 0, (self.patch_size, )*2).convert('RGB'))
if self.f is not None:
patch = self.f(patch)
return patch
# instantiate dataset with coordinates, img_path, desired patch_size, etc.
dataset = img_dataset(**kwargs)
# get the 2nd patch
second_patch = img_dataset[1]
# instantiate a dataloader
dataloader = torch.utils.data.Dataloader(dataset, num_workers=8, batch_size=32)
features = []
for batch_idx, imgs in enumerate(dataloader):
# do something (e.g. compute some summary statistics)
features.append(imgs.mean(axis=(0,1)))
closing due to inactivity