SnpSift
 SnpSift copied to clipboard
SnpSift copied to clipboard
Hi,i use the snpsift to annotate the vcf file ,it's very useful ;but i found that the same INFO/TAG in both anno.vcf and input.vcf,such as: a.vcf INFO= b.vcf INFO= i...
Hi, I have a 3.8Gb VCF file produced by a WGS pipeline mapping on hg38 and mutation calling using Mutect2. The file contains chromosomes chr1 ... chr22, chrX, chrY and...
The latest dbNSFP includes annotations for AlphaMissense and other things that did not exist before. However, if I download the latest dbNSFP, build it, and then use it to annotate:...
hadoop-bam and typesafe repo url were using http preventing maven from downloading sources
Bumps [htsjdk](https://github.com/samtools/htsjdk) from 2.24.1 to 3.0.1. Release notes Sourced from htsjdk's releases. 3.0.1 Fix for a long standing vulnerability around temporary directory creation which could expose data to malicious users...
After annating by snpsift, how culd I filter based on FREQ? For example, a record with the following annotation, and I would like to filter based on GnomAD. How should...
Hi, I have a vcf file annotated by snpeff and want to extract "ANN[*].RANK" from the ANN field like ANN=G|missense_variant|MODERATE|TNN|ENSG00000120332|transcript|ENST00000239462|protein_coding|13/19|c.3038C>G|p.Thr1013Arg|3151/5008|3038/3900|1013/1299|| java -jar /home/pmi/src/snpEff/SnpSift.jar extractFields my.vcf CHROM POS ID REF ALT...
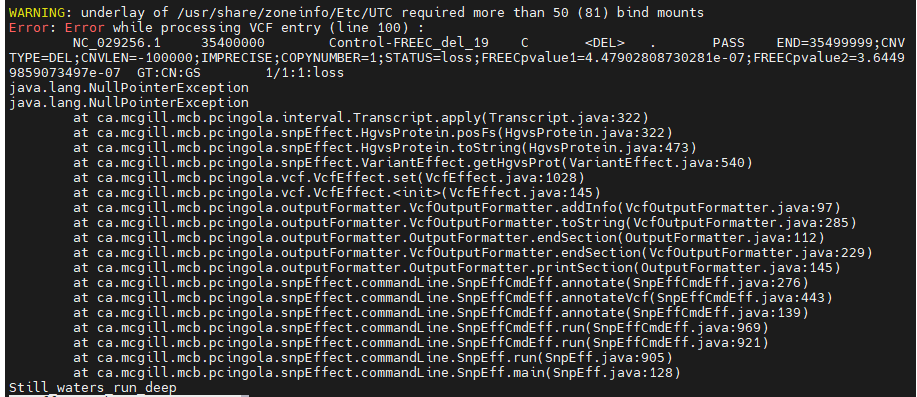 error while doing CNV anno
Hi developers, I'm using SnpSift to filter a VCF that has an entries for variants in a tumor sample and a corresponding normal sample. I'm trying to filter out any...
Thank you for SnpSift, what a powerful useful program! I'm new to SnpSift, and have a basic question - the VCF file I'm starting with already has an INFO.AF field....
