Pipelines do not work on ARM systems (e.g. Macbooks with M1/M2)
Have you checked the docs?
Description of the bug
The biocontainers images only support amd64 images (Intel/AMD CPUs) and thus, running pipelines on those machines will fail with an error similar to this one:
WARNING: The requested image's platform (linux/amd64) does not match the
detected host platform (linux/arm64/v8) and no specific platform was requested
Reason being that images are just build for amd64 and no option is given to also pick an ARM image:
$ manifest-tool inspect quay.io/biocontainers/blast:2.13.0--hf3cf87c_0
quay.io/biocontainers/blast:2.13.0--hf3cf87c_0: manifest type: application/vnd.docker.distribution.manifest.v2+json
Digest: sha256:221b0ab5540cf7c4013b51b60b2c66113104a5b700611d411ae25eb5904f78d8
Architecture: amd64
OS: linux
# Layers: 3
layer 1: digest = sha256:73349e34840e6f54750ec5df84f447c2a01df267a601af1ca0ee7dffed8715f3
layer 2: digest = sha256:acab339ca1e8ed7aefa2b4c271176a7787663685bf8759f5ce69b40e4bd7ef86
layer 3: digest = sha256:1a4ef5ea54c9a0c4cd84005215beddb4745c86663945d9ac7b11ea42f237cd1f
Since the quay.io/biocontainers repository is hard-coded in Nextflow it is hard to select different images from other repositories.
Blast-Example
This also applies to the blast-example.
$ docker run -ti --rm nextflow/examples
WARNING: The requested image's platform (linux/amd64) does not match the detected host platform (linux/arm64/v8) and no specific platform was requested
root@1b9d4420be97:/#
This blast command below just hangs forever, since the blastp command is an x86_64 binary.
$ file /opt/ncbi-blast-2.2.29+/bin/blastp
/opt/ncbi-blast-2.2.29+/bin/blastp: ELF 64-bit LSB executable, x86-64, version 1 (SYSV), dynamically linked (uses shared libs), for GNU/Linux 2.6.9, stripped
Command used and terminal output
$ ./nextflow run blast-example -with-docker
N E X T F L O W ~ version 22.04.5
Launching `https://github.com/nextflow-io/blast-example` [drunk_kalam] DSL2 - revision: c7e4282e26 [master]
executor > local (1)
[27/4b2770] process > blast (1) [ 0%] 0 of 1
[- ] process > extract -
Relevant files
No response
System information
Macbook Pro (Apple Silicon - M1 Max)
I think this is a bit more related to upstream things happening - e.g. bioconda not producing arm builds (as of now). There is a bit of discussion going on here: https://github.com/bioconda/bioconda-utils/issues/706 but no solution (at least for now?)
I reckon it is important to split the problem into two streams:
- Build Images
- Make images available
AFAIU the problem is such a hassle, because the community relies on biocontainers to build the image and make it available. Basically an end-to-end solution for the problem.
I'd like to tackle the problem in smaller chunks and allow for a solution that
- Creates images compatible with nf-core modules (ideally using bioconda, but not limited to it)
- Make those images available to everyone with an easy to use mirror config (either via nextflow config or via container runtime config)
- build up images as they are needed
I don't know if can be useful, but with Docker there is a way to emulate the x86_64 on M1... can this can be a transient solution?
@lucacozzuto potentially, but that might be dead slow to begin with and my idea allows to curate more images from the outside.
Yes, I was just thinking of a way to allow people to still use that architecture to run the pipelines and when the image is available they can just replace it
I did some benchmarking to get a sense of the impact of the workaround @snafees came up with. Her workaround adds --platform linux/amd64 which has Docker Desktop run in an emulated x86_64 environment.
Workaround
$ docker run -ti --rm --platform linux/amd64 alpine:3.16.2 uname -a
Linux 0cee9ab497ed 5.10.124-linuxkit #1 SMP PREEMPT Thu Jun 30 08:18:26 UTC 2022 x86_64 Linux
$ docker run -ti --rm alpine:3.16.1 uname -a
Linux 8e0a50976e29 5.10.124-linuxkit #1 SMP PREEMPT Thu Jun 30 08:18:26 UTC 2022 aarch64 Linux
When running emulated, one can observe the hypervisor qemu.
Benchmark
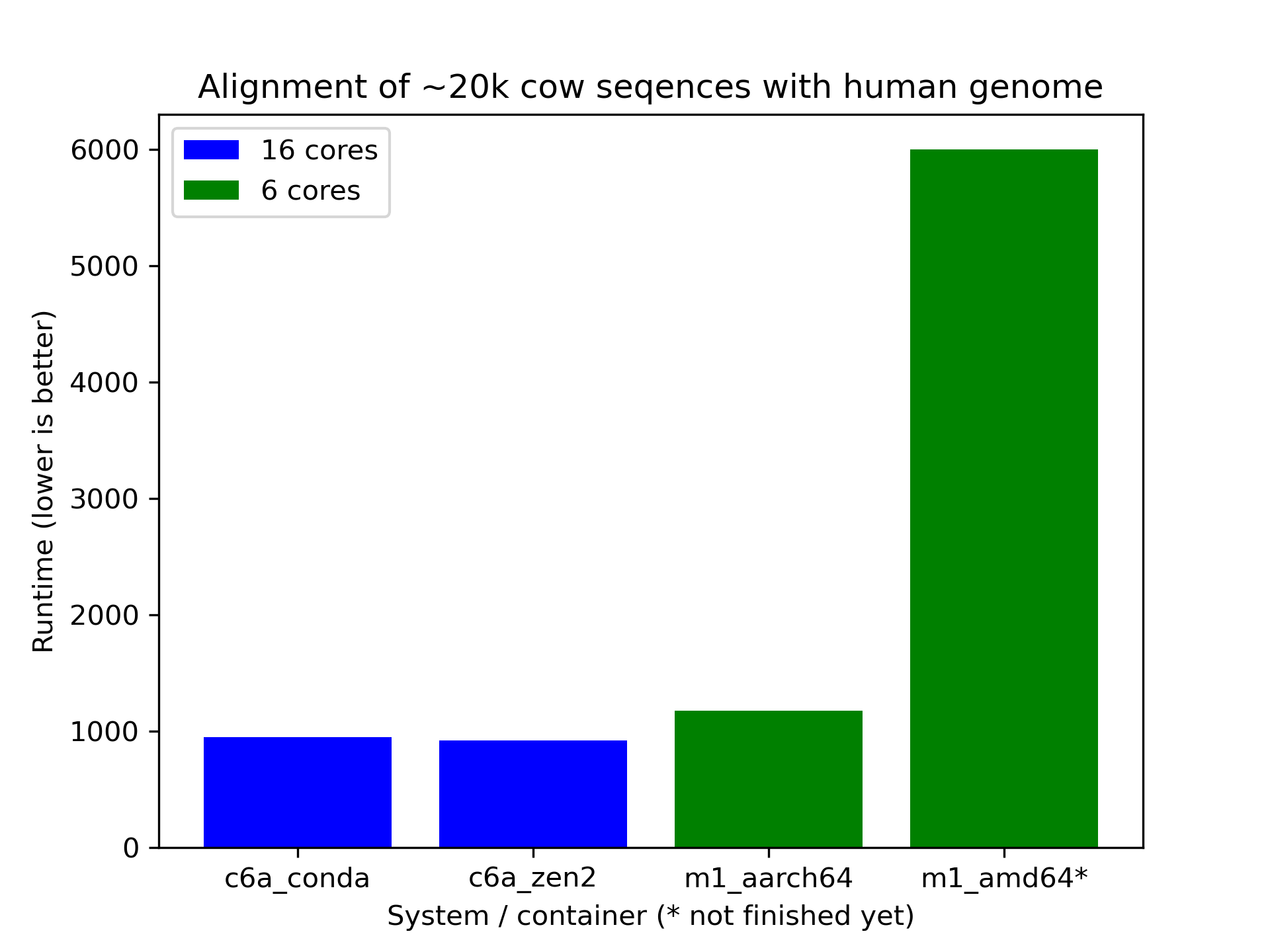
More data needs to be collected, but initial benchmarking indicates that the emulation has a significant impact on performance. I'll attach the notebook with the data down below.
# data (runtime of `blastp` in seconds)
m1_aarch64 = [1175]
m1_amd64 = [6000]
c6a_conda = [917, 984, 919, 963]
c6a_zen2 = [915, 930, 908, 923]
c7g_aarch64 = [799, 800, 797]
Runscript
cat > run-blast.sh << \EOF
#!/bin/bash
set -e
if [[ ! -f /blast-data/human.1.protein.faa ]];then
wget -qO /blast-data/human.1.protein.faa.gz ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot/human.1.protein.faa.gz
gunzip /blast-data/human.1.protein.faa.gz
fi
if [[ ! -f /blast-data/cow.1.protein.faa ]];then
wget -qO /blast-data/cow.1.protein.faa.gz ftp://ftp.ncbi.nih.gov/refseq/B_taurus/mRNA_Prot/cow.1.protein.faa.gz
gunzip /blast-data/cow.1.protein.faa.gz
fi
cp /blast-data/human.1.protein.faa human.1.protein.faa
head -200000 /blast-data/cow.1.protein.faa > cow.small.faa
echo "COW entries: $(grep -c '^>' cow.small.faa)"
makeblastdb -in human.1.protein.faa -dbtype prot
NPROCS=$(nproc)
start=$(date +%s)
set -x
time blastp -query cow.small.faa -db human.1.protein.faa -out results.txt -num_threads ${NPROCS}
set +x
end=$(date +%s)
runtime=$((end-start))
echo "Runtime ${NPROCS} tasks: ${runtime}"
EOF
chmod +x run-blast.sh
For each run we'll reuse a data volume which also includes the run script.
docker volume create blast-data2
docker run -ti --rm -v blast-data2:/blast-data -v $(pwd):/pwd alpine:latest cp /pwd/run-blast.sh /blast-data.sh
ARM64
docker run -ti --rm -v blast-data:/blast-data -v /scratch -w /scratch \
quay.io/cqnib/biocontainers-blast:2.13.0-aarch64 /blast-data/run-blast.sh
AMD64
docker run -ti --rm -v blast-data:/blast-data -v /scratch -w /scratch \
--platform=linux/amd64 \
quay.io/biocontainers/blast:2.13.0--hf3cf87c_0 /blast-data/run-blast.sh
An impact is expected in speed and is likely dependent on the tool as well. But is can be a temporary option for avoiding people with M1 cannot run anything. But I agree it should be just while new containers are available.
Yep, running slow is better than not running at all. And for python-scripts without a lot of interaction with the hypervisor that will be fine. But performance sensitive ones are going to suffer.
The --platform=linux/amd64 run is still not finished after 2h. I did not expect the impact to be that big.
Updated plot with Graviton2.
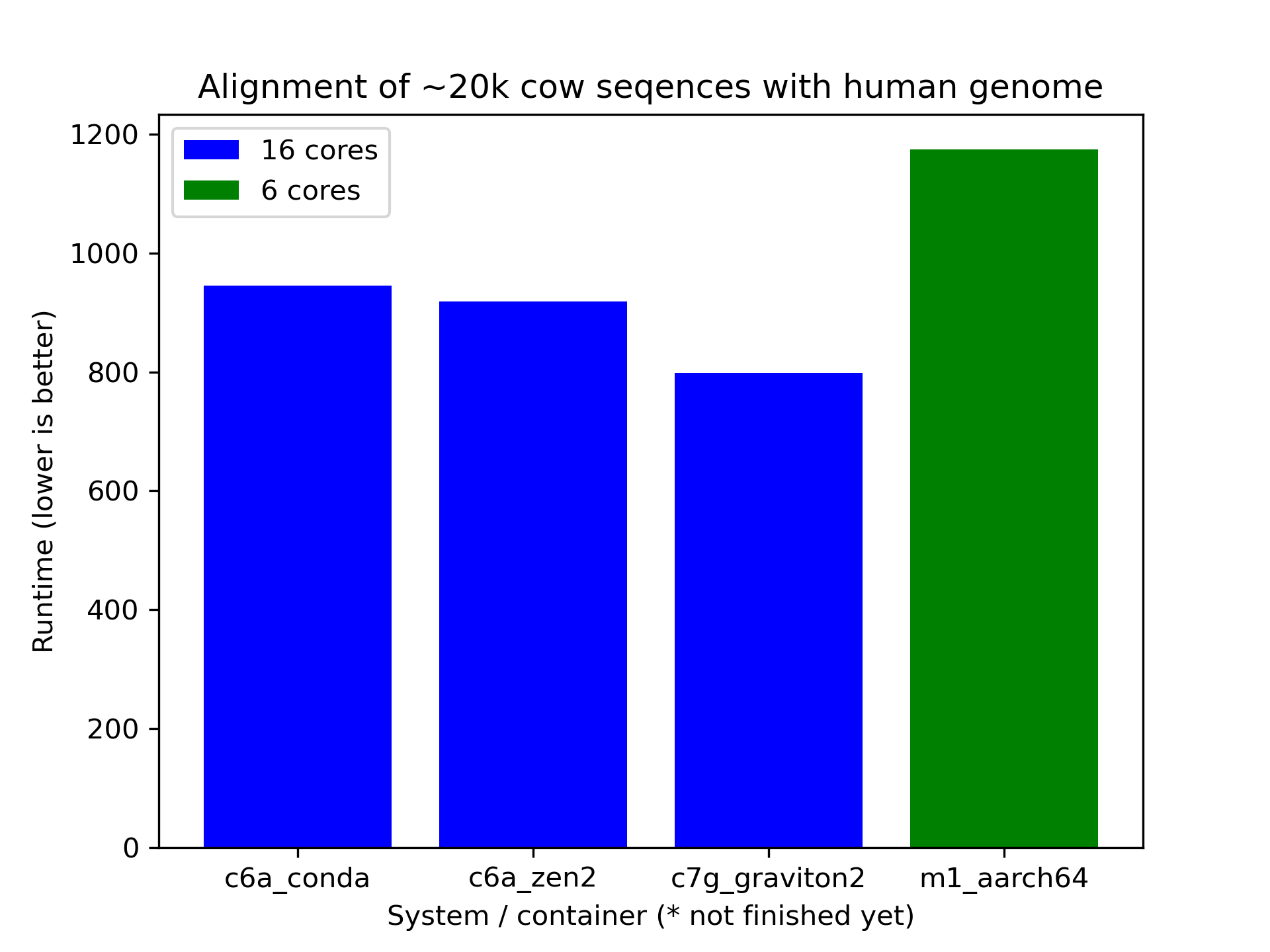
Conclusion is more benchmarking is required I suppose. :)
I also heard that you can use the GPU available on M1 macs...
https://towardsdatascience.com/installing-pytorch-on-apple-m1-chip-with-gpu-acceleration-3351dc44d67c