tombo
 tombo copied to clipboard
tombo copied to clipboard
Raise warning for empty reference sequences
Hello, I am using tombo to detect 5mC modification. (ubuntu server 18.04, ont-tombo==1.5, python==3.6, h5py==2.9.0)
The detect_modifications step failed with value error: no attribute name.
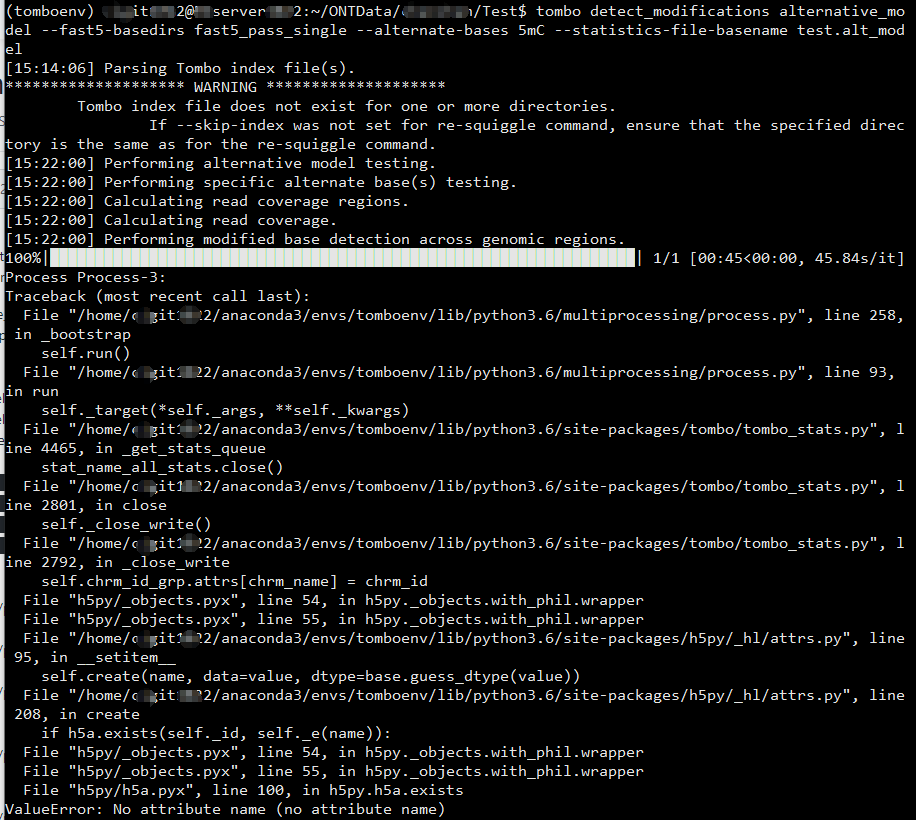
have checked the suggested solutions in the similar issue #115 ( leave off the per read statistics output ), but still got the error.
However, if I run the same processes on mac os, there is no errors.

Does that mean the current ont-tombo package is not compatible with the ubuntu linux?
Tombo is compatible with ubuntu/linux. Judging from the line numbers found in the error text it appears that the tombo version run on the ubuntu server is not the most recent code. Hopefully updating to the most recent github tombo version will resolve the hdf5 issues (e.g. install via pip install git+https://github.com/nanoporetech/tombo).
Thanks for the response. I will go check if updating to the most recent git version would resolve the issues.
reinstalled via https://github.com/nanoporetech/tombo.git.
still got the same issues. The line numbers in the error text changed though.
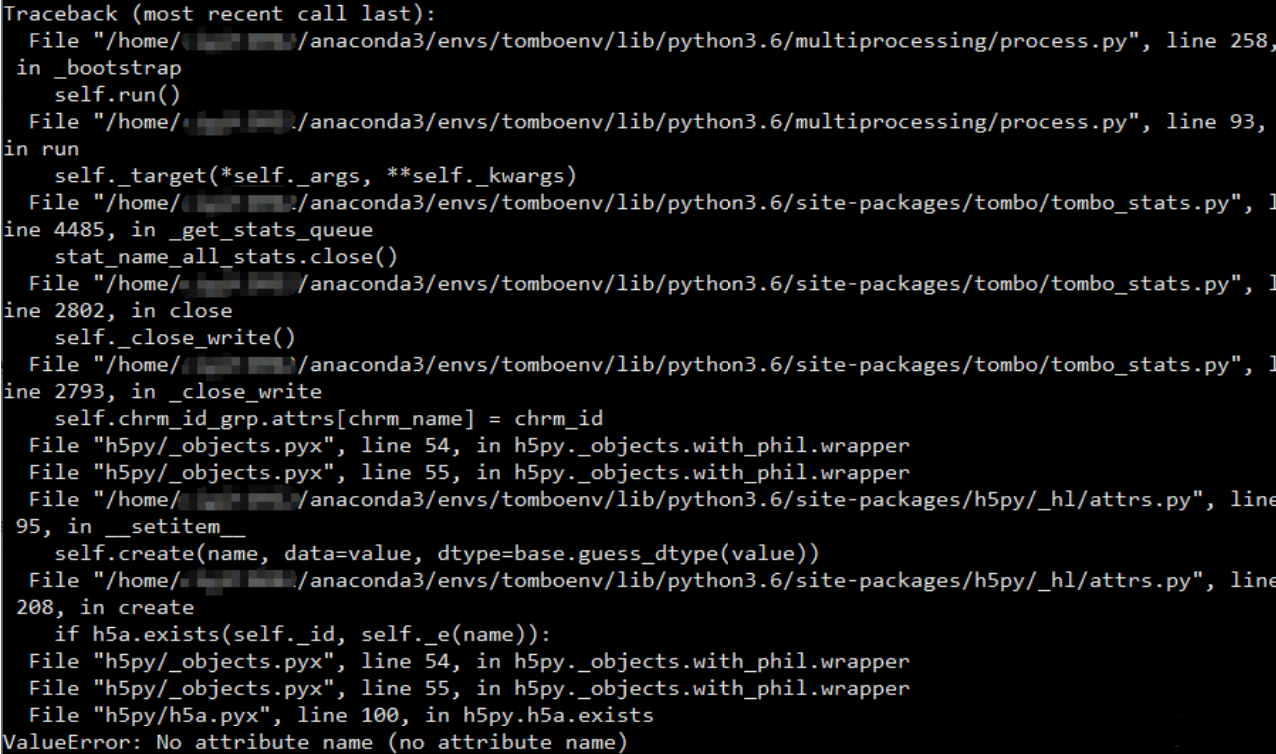
After few inspection, I found out that the error was caused by the empty sequence name in my manually created fasta files. The empty sequence name would cause that the chrm_name saved into the hidden index at re-squiggle step is empty, and produce the "no attribute name" error in the detect modification step.
Maybe it's better to replace the warning "[WARNING] empty sequence name in the input." in the re-squiggle step with an error reminding the user to check the fasta file.