Nico Matentzoglu
Nico Matentzoglu
Currently, some curie notations are clickable, like pmid: 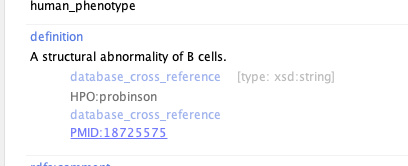 Clicking on PMID:18725575 brings me right to the paper. It would be super amazing if that would work for any kind...
- DOID:3666 is mapped to a string (cutaneous solitary mastocytoma) with skos:exactMatch, should be synonym - DOID:5546 is mapped to "cancer of the femur" with skos:exactMatch, should be synonym -...
We should document the new changes documentation in the developers pages, and think about how we can generalise this in the long run across all of OBO using KGCL and...
We use the following namespaces (aside from some spurious ones #2500) in Mondo. I am wondering whether we should use the official namespace (esp. SCTID, OMIM) for these. It seems...
Looking at Mondos equivalencies.owl, I found these: Being equivalent to Mondo classes. As there are no other (from any of these sources, like HP, Wikidata etc) classes equivalent to these,...
WARN when new proxy-merges occur (two classes of the same ontology get merged) Annotate all proxy merges (there may be some annotations) Query over reasoned graph ERROR: No MONDO class...
All sssom tables when run through split_sssom_by_source should be sorted entirely (like by all columns) and the columns should also be sorted in the sssom preferred order. Right now, some...
Currently, our skos content is a bit dirty: many duplicates. For example, these are the results when querying for skos:exactMatch: # OMIM duplicates: OMIM | MONDO -- | -- http://identifiers.org/omim/112500...
- [ ] @matentzn bring in corresponding genes for OMIM terms. Genes should be in the resulting RDF file - we should add the equiv axioms, where applicable
Also make sure better metadata (currently default).
