Adam Ginsburg
Adam Ginsburg
Minimalist WE: ```python from astropy.wcs import WCS from astropy.io import fits from astropy.visualization import simple_norm import pylab as pl from astropy import units as u hdul = fits.open('https://stpubdata-jwst.stsci.edu/ero/jw02731/L3/t/jw02731-o002_t017_miri_f1800w_i2d.fits') pl.figure(figsize=(10,10)) ww...
A [very frequent](https://github.com/search?q=proj_plane_pixel_scales&type=code) use pattern is, e.g.: ```python pixscale = wcs.utils.proj_plane_pixel_scales(cube_hi.wcs.celestial)[0] ``` in which the user: (1) assumes the pixels are square (2) wants only the linear transformation from pixel...
This documentation: http://docs.glueviz.org/en/stable/python_guide/glue_from_python.html#starting-glue-from-a-script tells you how to start from a script. It would be helpful to indicate what variables, if any, are predefined and which have to be created from...
Spectral axis type conversion (e.g., velocity frequency) should be available through the glue interface. A workaround using the terminal is: ```python data = dc[0] cube = data.get_object(cls=SpectralCube) data.coords = cube.with_spectral_unit(u.km/u.s,...
**Describe the bug** Creating a profile viewer then selecting a subset in the 2D viewer does not make the subset appear in the profile viewer **To Reproduce** ```python from glue.app.qt.application...
**Describe the bug** Exporting a session with data as relative paths includes the data **To Reproduce** See screenshot **Expected behavior** Smaller .glu flie! **Screenshots** 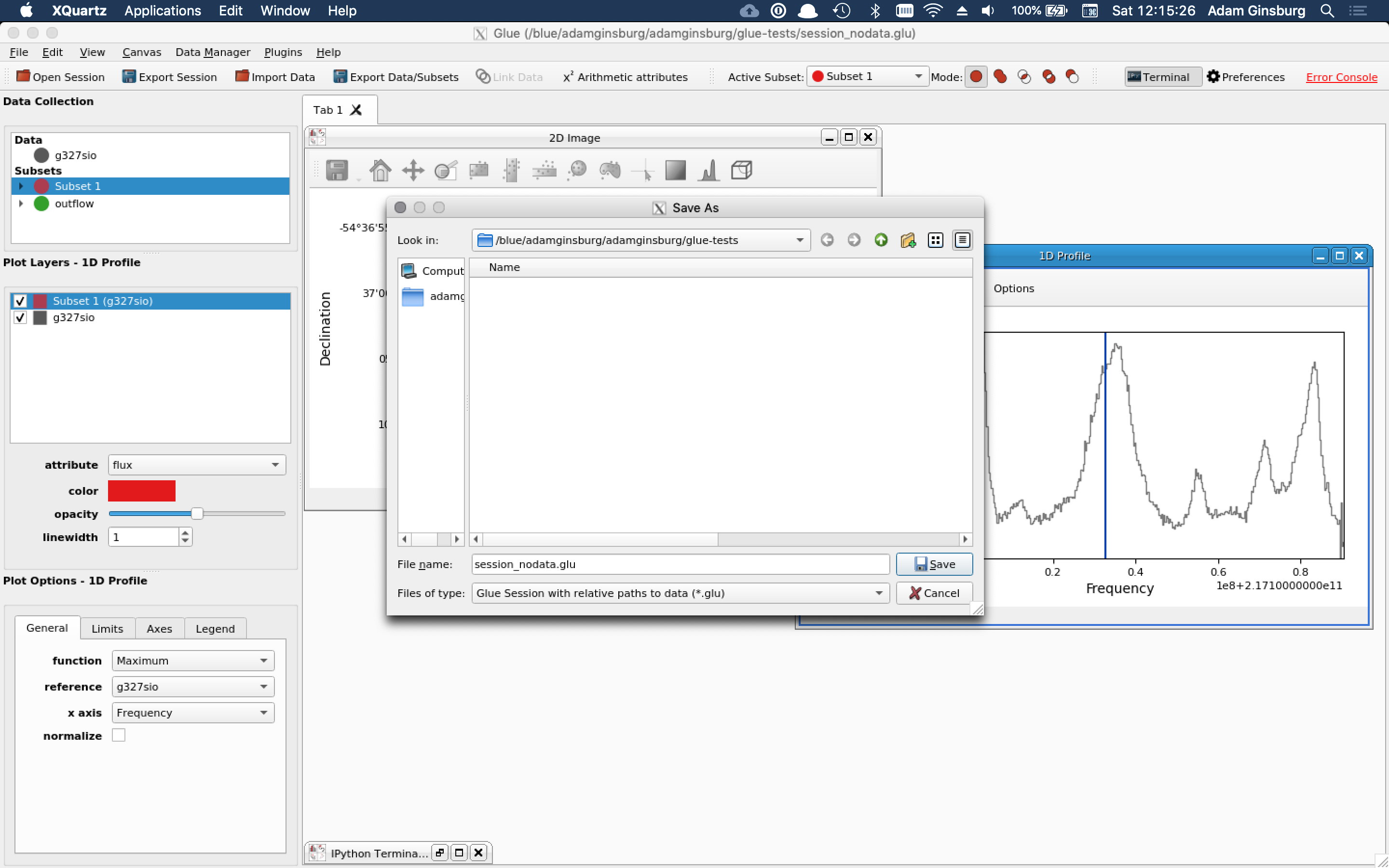 **Details:** ``` $ ls -lh...
Bally requested that a compass be displayable on images. In one of Alyssa's demo videos, one of the images is not aligned with the WCS axes (CROTA!=0), so it would...
This should fix #2489. The failure is already covered by tests.
The ALMA data downloader is broken because of an upstream change: ```python from astroquery.alma import Alma uid = 'uid://A001/X12a3/Xe9' data_info = alma.get_data_info(uid, expand_tarfiles=True) alma.retrieve_data_from_uid([uid]) ``` This fails on the second...
The current implementation only allows single isotopologues and molecules, which makes search for transitions difficult if the carrier species is unknown.
