UpSetR
 UpSetR copied to clipboard
UpSetR copied to clipboard
Stacked barplot for UpSetR ?
Hello,
Is it possible to combine stacked barplots based on different labels and UpSetR classes intersections in one plot ? I'm using a data frame where each item (row) is defined by: i) an intersection among the different sets (coded by 0 and 1) used by UpSetR ii) a text label that could be used by the stacked barplot
Using the example dataset "movies", the goal of this plot would be to analyse the number of movies released at each date for each intersection of genres.
I'm working with genomic data comprising regions of interest (one by row) belonging to various intersections among 7 sets (different conditions, correspondig to genres in the movies dataset) and each region is also defined by its location on the genome (genic, intergenic, promoter, corresponding to ReleaseDate in the movies dataset). I would thus be interested by combining UpSetR classes intersection representation with a stacked barplot that would take into account the localization of each item in the various genomic features (genic, intergenic, promoters).
I understand that such plot with many labels would be hard to interpret, but with only 3 labels for example, it could exhaustively represent my datasets in one plot.
Using the examples in the help of UpSetR, I did not see how to do it.
Any help would be greatly appreciated.
Yes, stacked barplots with attributes from other columns would be most helpful. There is a way to achieve 'pseudo-stacking' when active=T, but since the counts start from 0, interpretation becomes difficult. Also, forcing multiple queries to mimic stacking doesn't seem to work with the T or active=F option (with the triangles). An additional toggle parameter to make the counts cumulative could suffice for the time-being serving as a makeshift option.
Alternatively, stacked barplots from ggplot could be defined and added to attribute plots. But, these plots are made with the raw data and not the intersecting upset data (so, the axes per se wouldn't match with the bottom panel of the upset plot). Any thoughts on workarounds to use upset data for stacked plots? Thanks!
Below is an example of the incongruous plots that I've made to explain the question:

Also, not sure as to why the text and the set count bars don't align.
I love this idea - what would be very simple ( I THINK???) and useful would be to either make dodged bar plots, OR, to be able to overlay a point or a label on each bar. In my case I'm trying to show the "expected" overlap vs the "observed" overlap if that makes sense.
Yes, indeed, this would be very useful.
I have been trying to make a stacked barplot for a while, then realized that there is no "easy" way to create it. Since the issue has been raised for some time, was there any updates on this? It would be really nice if this could be added in the next version.
@jananiravi could you share the code that you use to build this stalked barplot on intersect area??
Is there any updated on this issue? I would be really interested in this feature.
@jananiravi @acpguedes I'm not really sure how to scale this up if you have more than two "lineages" as shown in @jananiravi's example. But here is my solution for a simpler case where each element in the set can either be UP or DOWN (for e.g. differentially expressed gene is up- or -down-regulated). The one "caveat" (or advantage, depending on your point of view) is that any given gene could be counted twice... once for being UP and a second time for being DOWN...


@jananiravi @acpguedes I'm not really sure how to scale this up if you have more than two "lineages" as shown in @jananiravi's example. But here is my solution for a simpler case where each element in the set can either be UP or DOWN (for e.g. differentially expressed gene is up- or -down-regulated). The one "caveat" (or advantage, depending on your point of view) is that any given gene could be counted twice... once for being UP and a second time for being DOWN...
@radlinsky, I am still unclear how you generate the stacked bar chart. Could you please share your code to reproduce your example?
Many thanks in advance!
Hey @janstrauss1, I thought I added a txt file with the code to reproduce the plot, but maybe it didn't attach. Here:
` library(UpSetR) test <- as.data.frame(matrix(rnorm(1:12), nrow = 4)) colnames(test) <- c("A", "B", "C") test_up <- test test_up[test >= 0] <- 1 test_up[test_up != 1] <- 0 test_up$Gene <- paste0("Gene", 1:nrow(test), "_UP") test_down <- test test_down[test < 0] <- 1 test_down[test_down != 1] <- 0 test_down$Gene <- paste0("Gene", 1:nrow(test), "_DOWN") test <- rbind(test_up, test_down)
upordown <- function(row, direction) { gene <- row["Gene"] if (grepl(x = gene, pattern = direction)) { newData <- T } else { newData <- F } }
metadata <- data.frame( c("A", "B", "C"), as.numeric(apply(test_up[, 1:3], 2, sum)) ) colnames(metadata) <- c( "sets", "NumberUP" )
upset(test, sets = c("A", "B", "C"), set.metadata = list( data = metadata, plots = list( list(type = "hist", column = "NumberUP", assign = 20, # defines width of the meta-data histogram colors = "red") ) ), queries = list(list(query = upordown, params = list("_UP"), color = "red", active = TRUE)) ) `
Sadly it looks like this project has been put on hold....
But here is my contribution to this possible stacked barplot feature to add to upsetr because i think it can be very useful to gather closely related informations on the same figure (for a scientific article).
Attached are the barplot and the upsetr plot I would have liked to combine easily and not using Inkscape manually to copy paste each barplot on top of the corresponding regroupments. Using this combined plot it is easier to understand and vizualize which samples share common feature and it can be easily be related to the composition of each.
1/ barplot alone

2/ upsetr plot alone
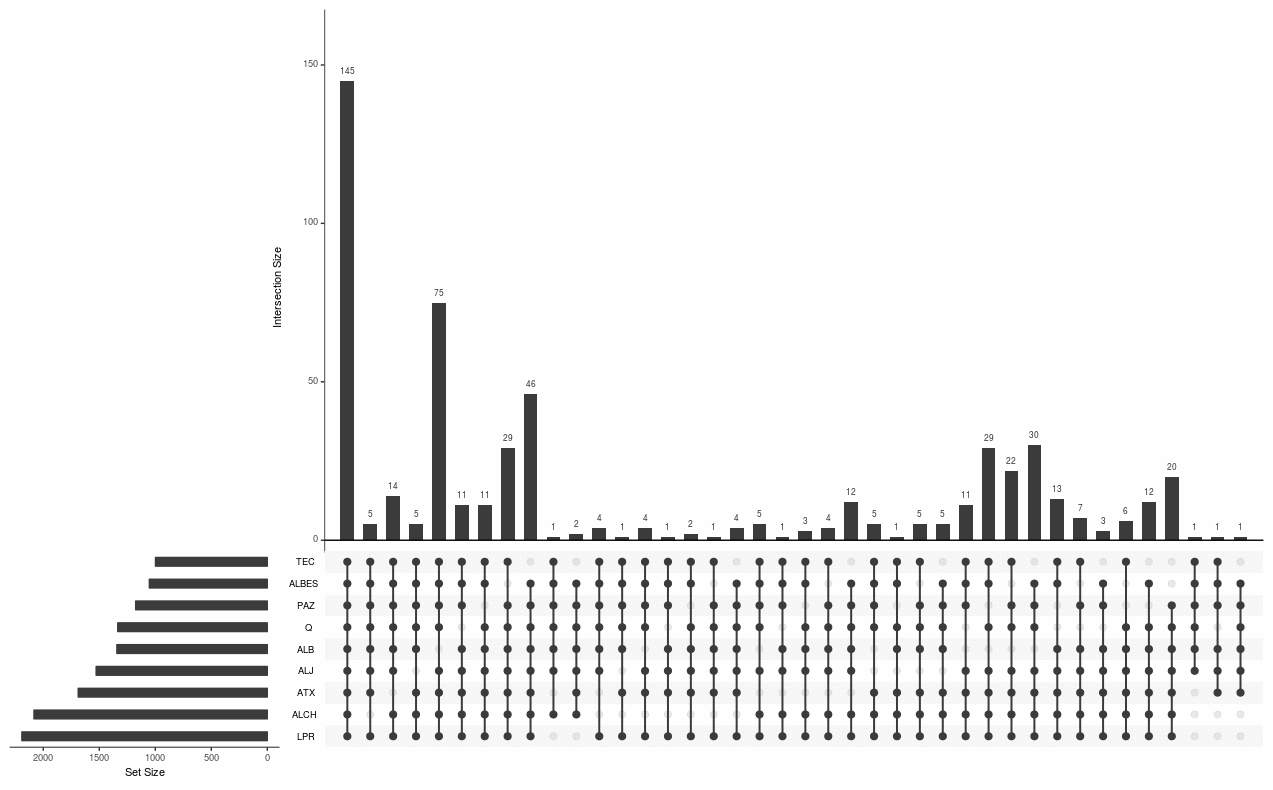
3/ both combined

Hope this does help to understand why it could be a really nice add-on to this package and what I (we) would expect to do then !
Thanks, Greboul
This question also arose on stackoverflow, and I found a workaround for the main (intersection) bars that I posted there: https://stackoverflow.com/a/56704255/2352071
What remains impossible with that workaround is overlaying the set sizes with the query annotation. This would be another nice feature to have.
- 1 for @dlaehnemann's suggestion - if this could be native it would be great. @dlaehnemann your stack answer is great - anyway to add a legend to show what the colours in the stacked barchart mean?
Sorry, I just resorted to creating that manually, as I only had a single plot.
But Example 5 in this tutorial might work:
https://cran.r-project.org/web/packages/UpSetR/vignettes/queries.html
Otherwise, there seems to be a recent reimplementation based on the grammar of graphics, that should allow for easier customization. Not sure if it supports as many panels in the grid, though: https://github.com/const-ae/ggupset
thanks those are both really helpful suggestions - Example 5 does indeed work.
I just saw this in my e-mail. I have a package which re-implements upset called ComplexUpset using ggplot2. Would the following work for you:

https://krassowski.github.io/complex-upset/articles/Examples_R.html#fill-the-bars
The code is slightly different but the gist is:
upset(
movies,
genres,
base_annotations=list(
'Intersection size'=intersection_size(
counts=FALSE,
aes=aes(fill=mpaa)
)
),
width_ratio=0.1
)
Nice, you might also want to add this info to the stackoverflow question, that does seem to get some traffic! :)
https://stackoverflow.com/a/56704255/2352071
@krassowski very neat, thanks
Very nice, I guess it closes the issue !