Francois Serra
Results
24
issues of
Francois Serra
make it use the bam file directly (but keeping the option to input matrix).
enhancement
convention
evaluate the need to do this with a separate tool, but tadbit tools would need to integrate: - comparison of compartements (plot + list of differences in bed format) -...
enhancement
plot distribution of interaction in ranges of: 20-40kb divided by 20-100kb. In this way chromosome size should not be source of bias. 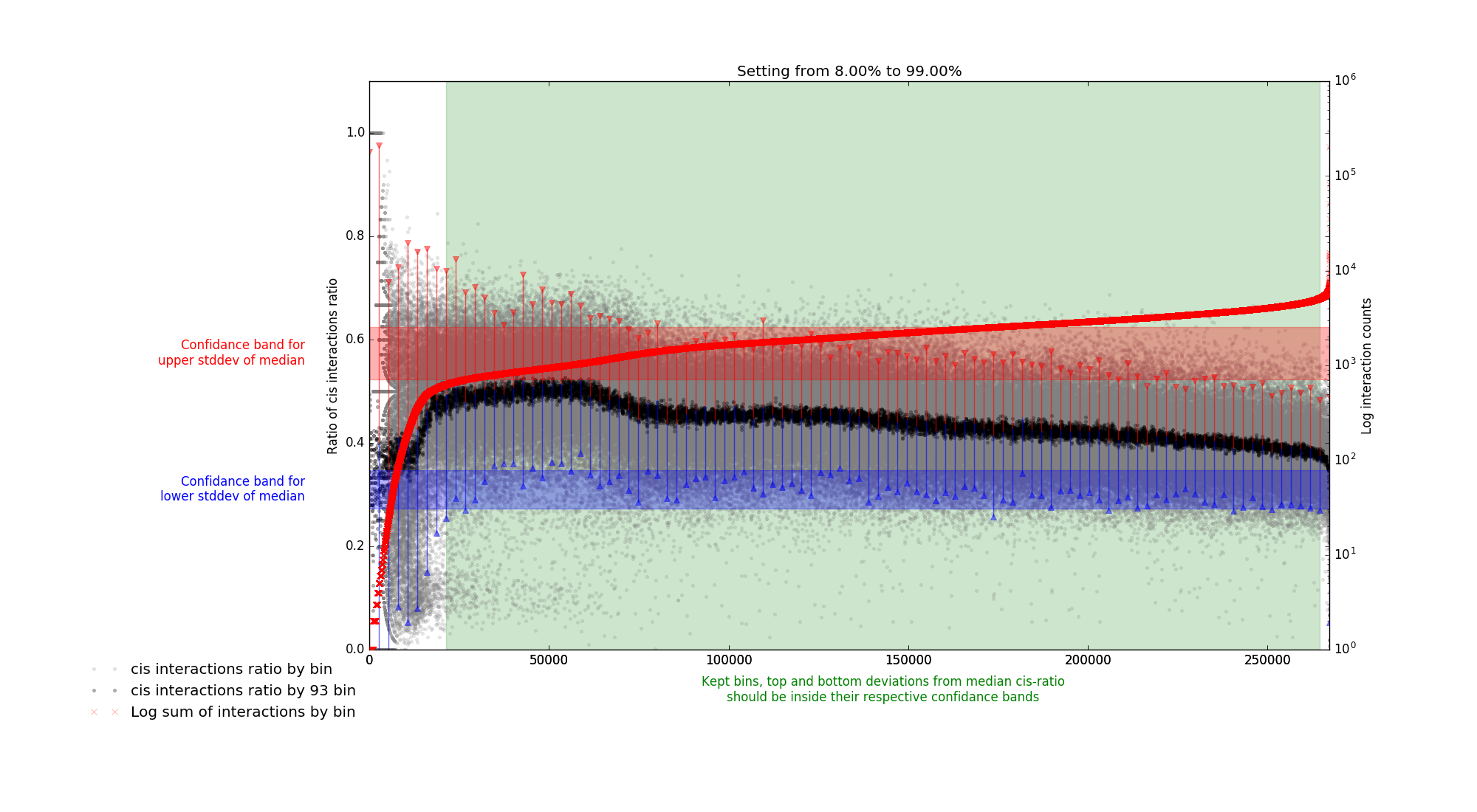
enhancement
IMPORTANT
convention
currently file type (fastq or map file) is assessed using the file name.
IMPORTANT
convention
@david-castillo found start and end of chromsomes may be wrong. add a flag to write matrix inside JSON file
bug
enhancement
in this case tad_height should be computed on RAW data
bug
convention
