Daniel Brennan
Daniel Brennan
@ntustison thanks for this insight. It makes sense that the SST would reveal thicker values given the info you've shared. Do you suggest the same as @cookpa and basically run...
sorry, what was the issue that was resolved? Single-subject longitudinal processing? I am unable to find the issue on github currently
Got it. Strangely, it seems that the “cross Sectional ” data is perfectly fit, with the longi data being under estimated. Still not sure why that’s happening. Anyways, what is...
@cookpa , you're correct other than X-axis is @jeffduda's pipeline and an older version of ANTs (I believe antsLongitudinalCorticalThickness.sh), y-axis is my custom pipeline which was made to emulate antsLongitudinalCorticalThickness...
@cookpa that makes sense  here is the "warpedPrior" brain mask (in lighter gray), with the posterior brain mask underneath. As you say, it inflates the mask to fill that...
 decent results using a 7th tissue prior edit: I had to subtract the 7th tissue prior from the GM tissue prior to achieve these results. the GM prior overwhelmed...
@cookpa it looks similar, in that it is estimating the entire gyrus as white matter
for example: 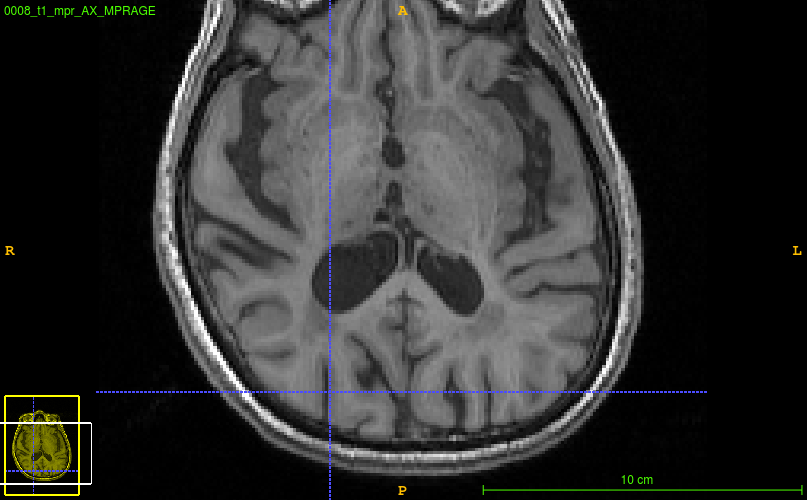 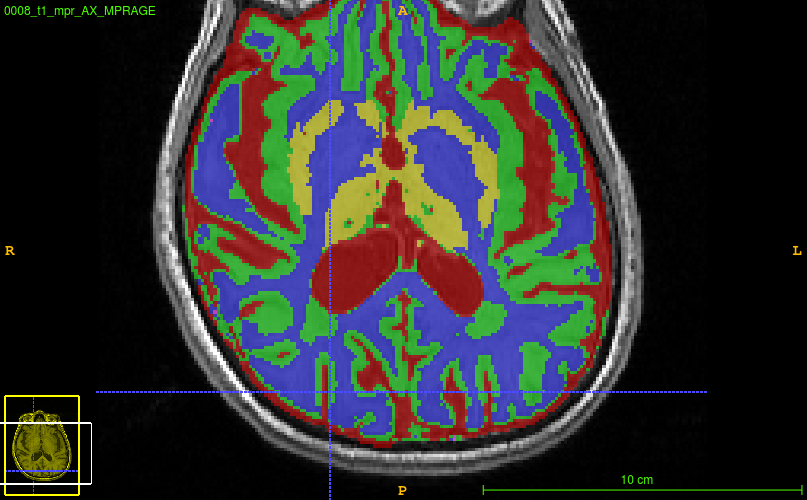 seems to be over-estimating white matter
@cookpa excellent suggestion, I will run and report results
should the prior weight be decreased or increased?