ntEdit
 ntEdit copied to clipboard
ntEdit copied to clipboard
✏️ Genome assembly polishing & SNV detection

ntEdit
Fast, lightweight, scalable genome sequence polishing & snv detection*
2018-2022
email: rwarren [at] bcgsc [dot] ca
*experimental feature
Description
ntEdit is a fast and scalable genomics application for polishing genome assembly drafts, with best performance on long read assemblies. It simplifies polishing, variant detection* and "haploidization" of gene and genome sequences with its re-usable Bloom filter design. We expect ntEdit to have additional application in fast mapping of simple nucleotide variations between any two individuals or species’ genomes.
! NOTE: In v1.3.1 onwards, the parameter k is automatically detected from supplied Bloom filters
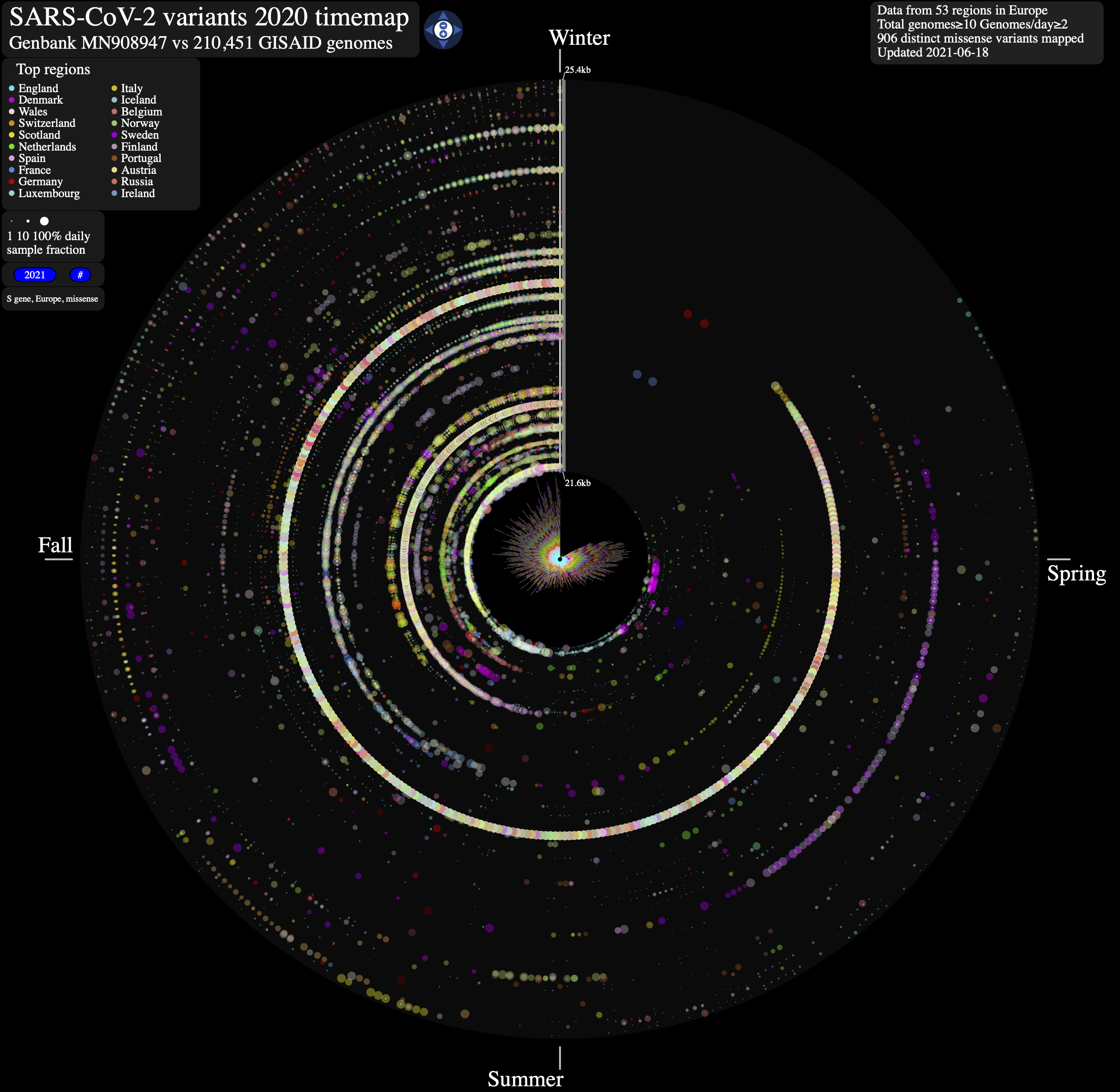
- SARS-CoV-2 evolution in human hosts. ntEdit v1.3.4 was used to map nucleotide variation between the first published coronavirus isolate from Wuhan in early January and over 1,500,000 SARS-CoV-2 genomes sampled from around the globe during the COVID-19 pandemic. Additional maps are available.
Implementation and requirements
ntEdit v1.2 and subsequent versions are written in C++.
(We compiled with gcc 5.5.0)
Install
Clone and enter the ntEdit directory.
https://github.com/bcgsc/ntEdit.git cd ntEdit
Compile ntEdit.
make ntedit
Dependencies
- ntHits (https://github.com/bcgsc/nthits)
- BloomFilter utilities (provided in ./lib)
- kseq (provided in ./lib)
Documentation
Refer to the README.md file on how to install and run ntEdit. Our manuscript contains information about the software and its performance. This ISMB2019 poster contains additional information, benchmarks and results.
Questions or comments? We would love to hear from you! [email protected]
Citing ntEdit
Thank you for your 
If you use ntEdit in your research, please cite:
ntEdit: scalable genome sequence polishing
ntEdit: scalable genome sequence polishing. Warren RL, Coombe L, Mohamadi H, Zhang J, Jaquish B, Isabel N, Jones SJM, Bousquet J, Bohlmann J, Birol I. Bioinformatics. 2019 Nov 1;35(21):4430-4432. doi: 10.1093/bioinformatics/btz400.
The experimental data described in our paper can be downloaded from: http://www.bcgsc.ca/downloads/btl/ntedit/
Credits
ntedit (concept, algorithm design and prototype): Rene Warren
nthits / nthash / BloomFilter.pm: Hamid Mohamadi
C++ implementation: Jessica Zhang, Rene Warren, Johnathan Wong
Integration tests: Murathan T Goktas
How to run in a pipeline
- Running nthits (please see nthits documentation : https://github.com/bcgsc/ntHits)
nthits -c-b -k -t reads eg. nthits -c 2 --outbloom -p solidBF -b 36 -k 25 -t 48 Sim_100_300_1.fq Sim_100_300_2.fq or nthits -c 2 --outbloom -p solidBF -b 36 -k 25 -t 48 @reads.in Where @reads.in is a file listing the path to all read fastq files to kmerize note the options --outbloom and -p must be set when using ntHits with ntEdit. If not specifying a hard threshold (-c), and relying instead on ntCard* to identify the error kmer coverage, please run with the --solid: nthits -b 36 -k 50 -t 48 --outbloom --solid @reads.in NOTE: THIS WILL WORK WELL WITH ntEdit ONLY IF YOU HAVE SUFFICIENT READ COVERAGE (>30X), OTHERWISE SET KMER COVERAGE TO -c2 (>=20X) or -c 1 (30X nthits -b 36 -k 40 -t 48 --outbloom --solid @reads.in Where @reads.in is a file listing the path to all read fastq files to kmerize -c sets a hard coverage threshold for reporting kmers (-c n : keeping kmers with coverage > n) --solid will output non-error kmers, as determined by ntCard. Use this option only when you don't wish to set the threshold (-c) manually. --outbloom simply outputs the coverage-thresholded kmers in a Bloom filter, whether it's from using -c or --solid. -b is the Bloom filter bit size. Use -b 36 to keep the Bloom filter false positive rate low (~0.0005). *About ntCard We recommend that you run ntCard independently on your short read data, and plot the kmer coverage distribution. Ideally, you may select a coverage threshold cutoff (nthits -c) based on the histogram. https://github.com/bcgsc/ntCard Bioinformatics. 2017 May 1; 33(9): 1324–1330. Published online 2017 Jan 5. doi: 10.1093/bioinformatics/btw832 PMCID: PMC5408799 PMID: 28453674 ntCard: a streaming algorithm for cardinality estimation in genomics data Hamid Mohamadi, Hamza Khan and Inanc Birol
- Running ntEdit (see complete usage below)
./ntedit -f-r -b -t eg. ./ntedit -f ecoliWithMismatches001Indels0001.fa -r solidBF_k25.bf -b ntEditEcolik25 -t 48
Running ntEdit
e.g. ./ntedit -f ecoliWithMismatches001Indels0001.fa -r solidBF_k25.bf -b ntEditEcolik25 ntEdit v1.3.5 Fast, lightweight, scalable genome sequence polishing & snv detection* Options: -t, number of threads [default=1] -f, draft genome assembly (FASTA, Multi-FASTA, and/or gzipped compatible), REQUIRED -r, Bloom filter file (generated from ntHits), REQUIRED -e, secondary Bloom filter with kmers to reject (generated from ntHits), OPTIONAL. EXPERIMENTAL -b, output file prefix, OPTIONAL -z, minimum contig length [default=100] -i, maximum number of insertion bases to try, range 0-5, [default=4] -d, maximum number of deletions bases to try, range 0-5, [default=5] -x, k/x ratio for the number of kmers that should be missing, [default=5.000] -y, k/y ratio for the number of editted kmers that should be present, [default=9.000] -X, ratio of number of kmers in the k subset that should be missing in order to attempt fix (higher=stringent), [default=0.5] -Y, ratio of number of kmers in the k subset that should be present to accept an edit (higher=stringent), [default=0.5] -c, cap for the number of base insertions that can be made at one position, [default=k*1.5] -j, controls size of kmer subset. When checking subset of kmers, check every jth kmer, [default=3] -m, mode of editing, range 0-2, [default=0] 0: best substitution, or first good indel 1: best substitution, or best indel 2: best edit overall (suggestion that you reduce i and d for performance) -s, SNV mode. Overrides draft kmer checks, forcing reassessment at each position (-s 1 = yes, default = 0, no. EXPERIMENTAL) -a, Soft masks missing kmer positions having no fix (-v 1 = yes, default = 0, no) -v, verbose mode (-v 1 = yes, default = 0, no) --help, display this message and exit --version, output version information and exit If one of X/Y is set, ntEdit will use those parameters instead. Otherwise, it uses x/y by default.
ntEdit Modes
Mode 0: (default) ntEdit will try to substitute the last base of an incorrect k-mer with a different ATGC base. If that k-mer is found in the bloom filter and has enough subset support, ntEdit will then try the other substitution bases and then choose the best substitution fix. However, if the substituion was not found, then ntEdit will try all indels of max length (-i) and (-d) starting with that substitution base and make edit based on the first accepted indel. Mode 1: ntEdit will try to substitute the last base of an incorrect k-mer with a different ATGC base. If that k-mer is found in the bloom filter and has enough subset support, ntEdit will then try the other substitution bases and then choose the best substitution fix. However, if the substitution was nto found,t hen ntEdit will try all indels of max length (-i) and (-d) starting witht hat substitution base and make edit based on the best accepted indel. Mode 2: ntEdit will choose the best substitution or indel for each incorrect k-mer. Since this can be very computationally expensive because ntEdit tries every combination possible, it is recommended that you reduce (-i) and (-d).
*We recommend running ntEdit in Mode 1 (or 0)
ntEdit -s (SNV) EXPERIMENTAL option
Version 1.3 implements a new mode (-s 1) to help detect simple base variation in genome sequences. It works by overriding the kmer absence verification stage of ntEdit, effectively testing every base position for possible alternate k kmers. At the moment, ntEdit only reports possible base substitutions (no indels), along with the number of supported kmers (the latter is NOT a proxy for read/kmer coverage). In our tests on simulated (C. elegans, H. sapiens) and experimental (GIAB, HG001/HG004), we find k52/k55 (-j 3 -- see below) to give the best performance. Caveats: Variations occurring within 2*k are not reported. Because kmers are shorter and have less sequence context than reads and read pairs, kmer variations that occur within a genomic allele (intra allelic) may be reported. In order to minimize false discovery, we recommend using a secondary Bloom filter built with repeat kmers (see details on the -e Secondary Bloom filter option below). This option is provided as a convenience feature, implemented to do a quick and dirty variation detection analysis on large genomes. It is a basic presence/absence detector based on kmer subsets. For robust variant identification, we recommend statistically principled approaches. VCF output (v1.3.2+ _variants.vcf): We assume a diploid genome for reporting on the possible genotype (GT). Users working on polyploid genomes should chose to ignore the last two columns of the VCF file (ie. FORMAT INTEGRATION)
ntEdit -e Secondary Bloom filter, with kmers to exclude. EXPERIMENTAL option
A secondary Bloom filter with kmers to exclude may be provided to ntEdit v1.3 with the -e option. This option is useful when running ntEdit in -s 1 mode, effectively taking a "slice" of robust (non-error and non-repeated) kmers. For example: -f draft from A (pseudo haploid) -r Bloom filter from A (diploid): non-error kmers -e Bloom filter from A (diploid): repeat kmers -s 1 When computing a repeat kmer Bloom filter with ntHits, we recommend that you first run ntCard and plot the kmer coverage histogram to identify the repeat cutoff value. A higher precision can be achieved with a more strict repeat filter cutoff, albeit at the risk of impacting sensitivity. Alternate kmers that pass the presence verification stage of ntEdit will not be considered if present in the secondary Bloom filter. This option may be used to map homozygous variation between species/individual. For example, in such a set up: -f draft from A (pseudo haploid) -r Bloom filter from B (different individual or species, diploid): non-error kmers -e Bloom filter from A (diploid): non-error kmers -s 0 This will map sites that are different (homozygous variants) between B and A. *These are provided as examples. Other experimental setup are possible. We recommend setting the jump parameter (j) to 1 when using the secondary Bloom filter or using the following compatible j/k values: -j 1: all k combinations may be used -j 2: k31, k33, k35, k37, k39, k41, k43, k45, k47, k49, k51, k53, k55, k57, k59, k61 (odd k value) -j 3: k31, k34, k37, k40, k43, k46, k49, k52, k55, k58, k61 Faster ntEdit runs are achieved at -j 3. Higher values of j would not provide enough kmers in the k subset and have not been tested. Users should perform their own benchmarks.
ntedit-make
The ntEdit pipeline can be run with a Makefile (ntedit-make), which will run ntHits, then ntEdit for you.
- Inputs: Read file(s), draft genome to be polished.
- Outputs: Polished scaffolds in FASTA format.
- If you submit more than one read file, ensure each file has the same prefix (ex. myReads1.fq.gz, myReads2.fq.gz, ...) and specify the prefix with "reads=prefix". Ensure each read file is in an acceptable format (fastq, fasta, gz, bz, zip).
- We suggest either specifying the cutoff parameter or defining solid=true in your command to set it automatically.
Example usage:
- Draft genome to be polished: myDraft.fa
- Input reads: myReads1.fq, myReads2.fq
./ntedit-make ntedit draft=myDraft.fa reads=myReads k=64 cutoff=2 or ./ntedit-make ntedit draft=myDraft.fa reads=myReads k=64 solid=true
For more information about usage:
./ntedit-make help
Test data
Go to ./demo
(cd demo)
run:
./runme.sh
(../ntedit -f ecoliWithMismatches001Indels0001.fa.gz -r solidBF_k25.bf -d 5 -i 4 -b ntEditEcolik25)
ntEdit will polish an E. coli genome sequence with substitution error ~0.001 and indels ~0.0001 using pre-made ntHits Bloom filter
Expected files will be: ntEditEcolik25_changes.tsv ntEditEcolik25_edited.fa
Compare with: nteditk25_changes.tsv nteditk25_edited.fa
How it works
 Sequence reads are first shredded into kmers using ntHits, keeping track of kmer multiplicity. The kmers that pass coverage thresholds (ntHits, -c option builds a filter with kmers having a coverage higher than c) are used to construct a Bloom filter (BF). The draft assembly is supplied to ntEdit (-f option, fasta file), along with the BF (-r option) and sequences are read sequentially. Sequence strings are shredded into words of length k (kmers) at a specified value (-k option in versions before v1.3.1. In newer releases, k is detected automatically from the main Bloom filter supplied in -r) matching that used to build the BF, and each kmer from 5’ to 3’ queries the BF data structure for presence/absence (step 1). When a kmer is not found in the filter, a subset (Sk) of overlapping k kmers (defined by k over three, k/3) containing the 3’-end base is queried for absence (step 2). The subset Sk, representing a subsampling of k kmers obtained by sliding over 3 bases at a time over k bases, is chosen to minimize the number of checks against the Bloom filter. Of this subset, when the number of absent (-) kmers matches or exceeds a threshold defined by Sk- >= k/x (-x option), representing the majority of kmers in Sk, editing takes place (step 3 and beyond), otherwise step 1 resumes. In the former case, the 3’-end base is permuted to one of the three alternate bases (step 3), and the subset (Sk_alt) containing the change is assessed for Bloom filter presence (+). When that number matches or exceeds the threshold defined by Sk_alt+ >= k/y (-y option), which means the base substitution qualifies, it is tracked along with the number of supported kmers and the remaining alternate 3’-end base substitutions are also assessed (ie. resuming step 3 until all bases inspected). If the edit does not qualify, then a cycle of base insertion(s) and deletion(s) of up to –i and –d bases begins (step 4, -i option and step 5, -d option, respectively). As is the case for the substitutions, a subset of k kmers containing the indel change is tested for presence. If there are no qualifying changes, then the next alternate 3’-end base is inspected as per above; otherwise the change is applied to the sequence string and the next assembly kmer is inspected (step 1). The process is repeated until a qualifying change or until no suitable edits are found. In the latter case, we go back to step 1. When a change is made, the position on the new sequence is tracked, along with an alternate base with lesser or equal k kmer subset support, when applicable. Currently, ntEdit only tracks cases when edits are made (steps 3-5), and does not flag unedited, missing draft kmers (steps 1-2).
Sequence reads are first shredded into kmers using ntHits, keeping track of kmer multiplicity. The kmers that pass coverage thresholds (ntHits, -c option builds a filter with kmers having a coverage higher than c) are used to construct a Bloom filter (BF). The draft assembly is supplied to ntEdit (-f option, fasta file), along with the BF (-r option) and sequences are read sequentially. Sequence strings are shredded into words of length k (kmers) at a specified value (-k option in versions before v1.3.1. In newer releases, k is detected automatically from the main Bloom filter supplied in -r) matching that used to build the BF, and each kmer from 5’ to 3’ queries the BF data structure for presence/absence (step 1). When a kmer is not found in the filter, a subset (Sk) of overlapping k kmers (defined by k over three, k/3) containing the 3’-end base is queried for absence (step 2). The subset Sk, representing a subsampling of k kmers obtained by sliding over 3 bases at a time over k bases, is chosen to minimize the number of checks against the Bloom filter. Of this subset, when the number of absent (-) kmers matches or exceeds a threshold defined by Sk- >= k/x (-x option), representing the majority of kmers in Sk, editing takes place (step 3 and beyond), otherwise step 1 resumes. In the former case, the 3’-end base is permuted to one of the three alternate bases (step 3), and the subset (Sk_alt) containing the change is assessed for Bloom filter presence (+). When that number matches or exceeds the threshold defined by Sk_alt+ >= k/y (-y option), which means the base substitution qualifies, it is tracked along with the number of supported kmers and the remaining alternate 3’-end base substitutions are also assessed (ie. resuming step 3 until all bases inspected). If the edit does not qualify, then a cycle of base insertion(s) and deletion(s) of up to –i and –d bases begins (step 4, -i option and step 5, -d option, respectively). As is the case for the substitutions, a subset of k kmers containing the indel change is tested for presence. If there are no qualifying changes, then the next alternate 3’-end base is inspected as per above; otherwise the change is applied to the sequence string and the next assembly kmer is inspected (step 1). The process is repeated until a qualifying change or until no suitable edits are found. In the latter case, we go back to step 1. When a change is made, the position on the new sequence is tracked, along with an alternate base with lesser or equal k kmer subset support, when applicable. Currently, ntEdit only tracks cases when edits are made (steps 3-5), and does not flag unedited, missing draft kmers (steps 1-2).
OUTPUT FILES
| Output files | Description |
|---|---|
| _changes.tsv | tab-separated file; ID bpPosition+1 OriginalBase NewBase Support k-mers (out of k/j) AlternateNewBase Alt.Support k-mers eg. U00096.3_MG1655_k12 117 A T 9 |
| _edited.fa | fasta file; contains the polished genome assembly |
| _variants.vcf | vcf file; contains variant calls |
License
ntEdit Copyright (c) 2018-2022 British Columbia Cancer Agency Branch. All rights reserved.
ntEdit is released under the GNU General Public License v3
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, version 3.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.
For commercial licensing options, please contact Patrick Rebstein [email protected]




