ahwanpandey
ahwanpandey
Yes I wanted to do the same as well. Are there any plans to implement this? Thanks.
Hi Again, It would be great if I could get some advice on this! Best.
Thanks so much! This tool is really awesome! I had been doing a lot of what this tool does ad hoc using bits and pieces of code but to have...
Also, we are investigating frequently mutated pathways in say Ovarian Cancer and Homologous Recombination Deficiency (HRD) genes and it would make perfect sense to just view the mutation burden in...
Hey thanks for this! I am away until early next week but I will test as soon as I am back.
Looks like it's working. Thanks! 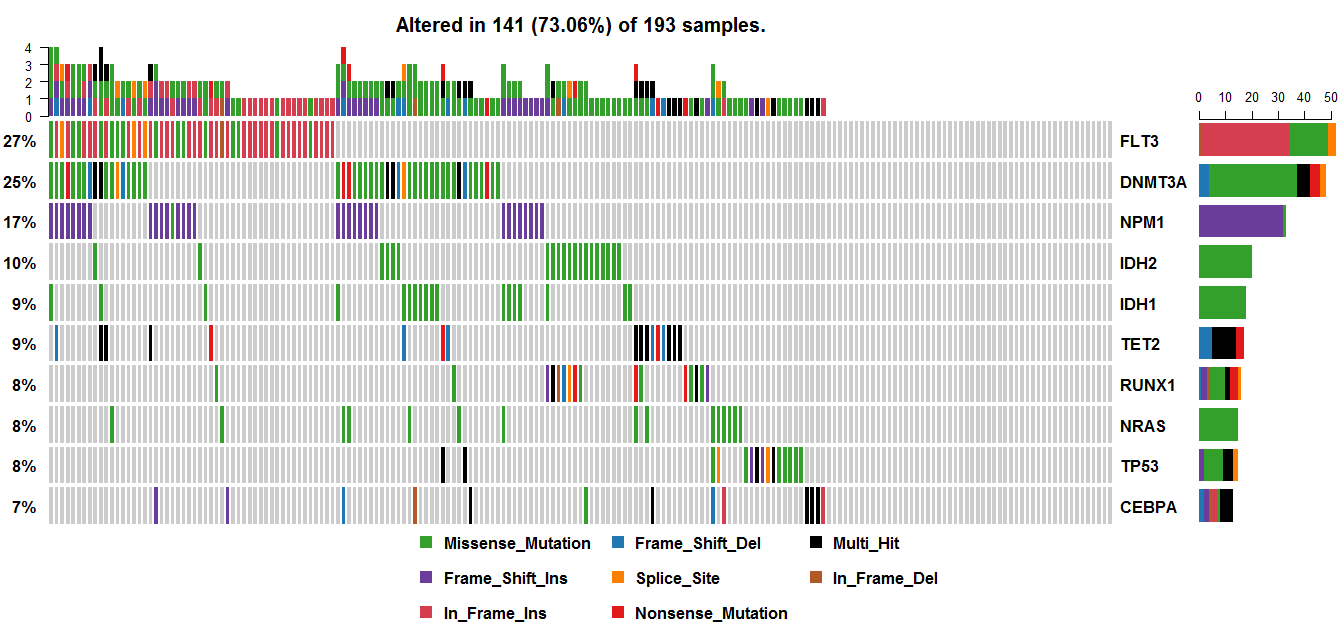
Hi, looks like the latest version of maftools has revertedd back to showing the "total mutations" in the column barplot rather than the mutations in the current pathway? Can the...
I am having the same issue with `biocontainers/biocontainers:v1.2.0_cv1` and `google/deepvariant:1.4.0`. Please help. ``` $ nf-core download \ > nanoseq --revision 3.1.0 \ > --outdir nf-core-nanoseq \ > --container-system singularity \...
OK managed to get the `google/deepvariant:1.4.0` container using the command parameters `-l quay.io -l ghcr.io -l docker.io`. ``` nf-core download \ nanoseq --revision 3.1.0 \ --outdir nf-core-nanoseq \ --container-system singularity...
I am having a similar issue. The error happens from chromosome 13 onwards. This is in windows ``` library(Rsamtools) > fastaFile FASTA FASTA class: FaFile path: F:/ahwan.pandey/Projects/TargetedExome/Project_DG/MOCOG/ref_test/human_g1k_v37.fasta index: F:/ahwan.pandey/Projects/TargetedExome/Project_DG/MOCOG/ref_test/human_g1k_v37.fasta.fai gzindex:...