Moss Voyager
Moss Voyager
Creating directory tmp_jkulsosc/. Error in file /users/gaoxl/soft/miniconda3/bin/fix_in_frame_stop_codon_genes.py at line 225: /users/gaoxl/soft/Augustus-3.3.1-tag1/scripts/gtf2gff.pl is not executable!
电脑端没问题 手机端搜不到 搜到了也播不了
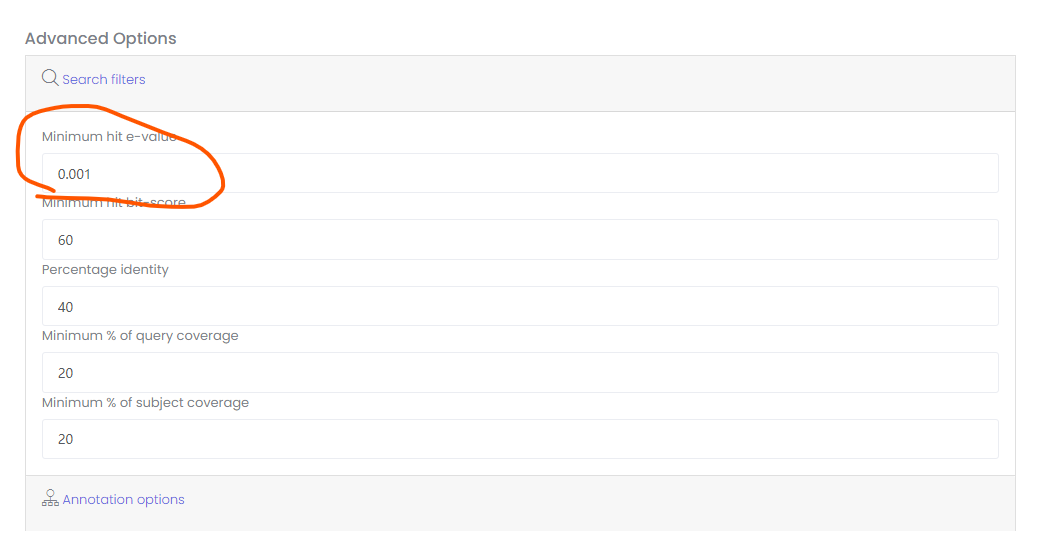 Hello, thank you very much for writing this software, this software has helped me a lot. But I encountered a problem when using the web version. The value parameter...
每次复制单独的对话内容时候,都会在末尾附带分割线代码,请问这个去掉是不是好一些? 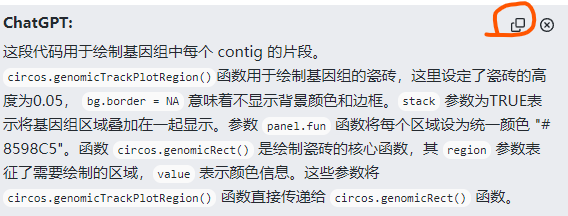 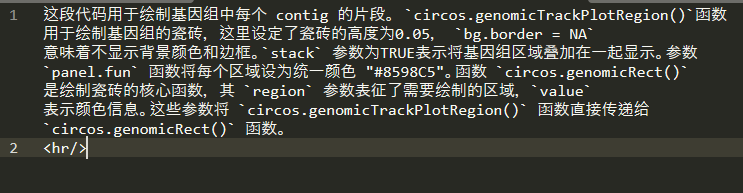
Hello, I have encountered some problems when I run Braker, the running process stops, but no error is reported. Here is my command: nohup /users/gaoxl/soft/miniconda3/bin/braker.pl --cores=8 --genome /users/gaoxl/work/7small_peptide_prediction_in_tomato/braker2/S_lycopersicum_chromosomes.4.00.fa --softmasking --bam...
Hello, there is no CDS information in the file I generated using gffcompare. But I want to continue to use gffread to generate CDS and protein sequences on this basis....
Hello, the genome (27m) I assembled using the standard process is 10m smaller than before using the software, but the genome size of this species is indeed the original genome...
Hello, when I ran the following command, I got a very large log file: 2.log, below is a screenshot of its content (not intercepted is a regular repetition) Run command:nohup...
