fix: mark MAR08430 as spontaneous
Main improvements in this PR:
This PR resolves #410.
I hereby confirm that I have:
- [x] Selected
developas a target branch
looks good to me
Thanks @JonathanRob. In addition to doing that, I was also intending to add the reference used in the Wikipedia page to MAR08430.
Sorry for eagerly marking MAR09927 without looking at the compartments.
For the intended reaction MAR08430 I am having a hard time finding support for the diffusion in the cited article PMID:3529851. My suggestion would be, prior to merging, for this to be checked.
Sorry for the mess, I wonder if I have not misunderstood this althogether. I think what they meant by the diffusion is that it diffuses to the fibrils from OUTSIDE mitochondria, so this is not related to actually passing through the membrane. Reading more on https://en.wikipedia.org/wiki/Creatine_kinase supports this - it seems that there are something called PCr/Cr-shuttle that facilitate this. There should probably be a GPR here, but I didn't find anything when googling, so maybe we should just stop this and not assign anything as spontanous. If anyone has an idea of how to figure out the GPR, it would be nice though. Again, sorry for the mess.
it seems that there are something called PCr/Cr-shuttle that facilitate this. There should probably be a GPR here, but I didn't find anything when googling, so maybe we should just stop this and not assign anything as spontaneous.
~~No solid diffusion evidence found by browsing the citations listed in the wiki page of Creatine phosphate shuttle. Such shuttles usually should get involved with transporters, i.e. with GPR.~~
According to this reported schematic view of a creatine-creatine phosphate shuttle, rephosphorylation of creatine is not occurred in mitochondrial matrix [m] but within intermembrane space [i], where both creatine and creatine phosphate can diffuse in and out through the highly permeable outer membrane.
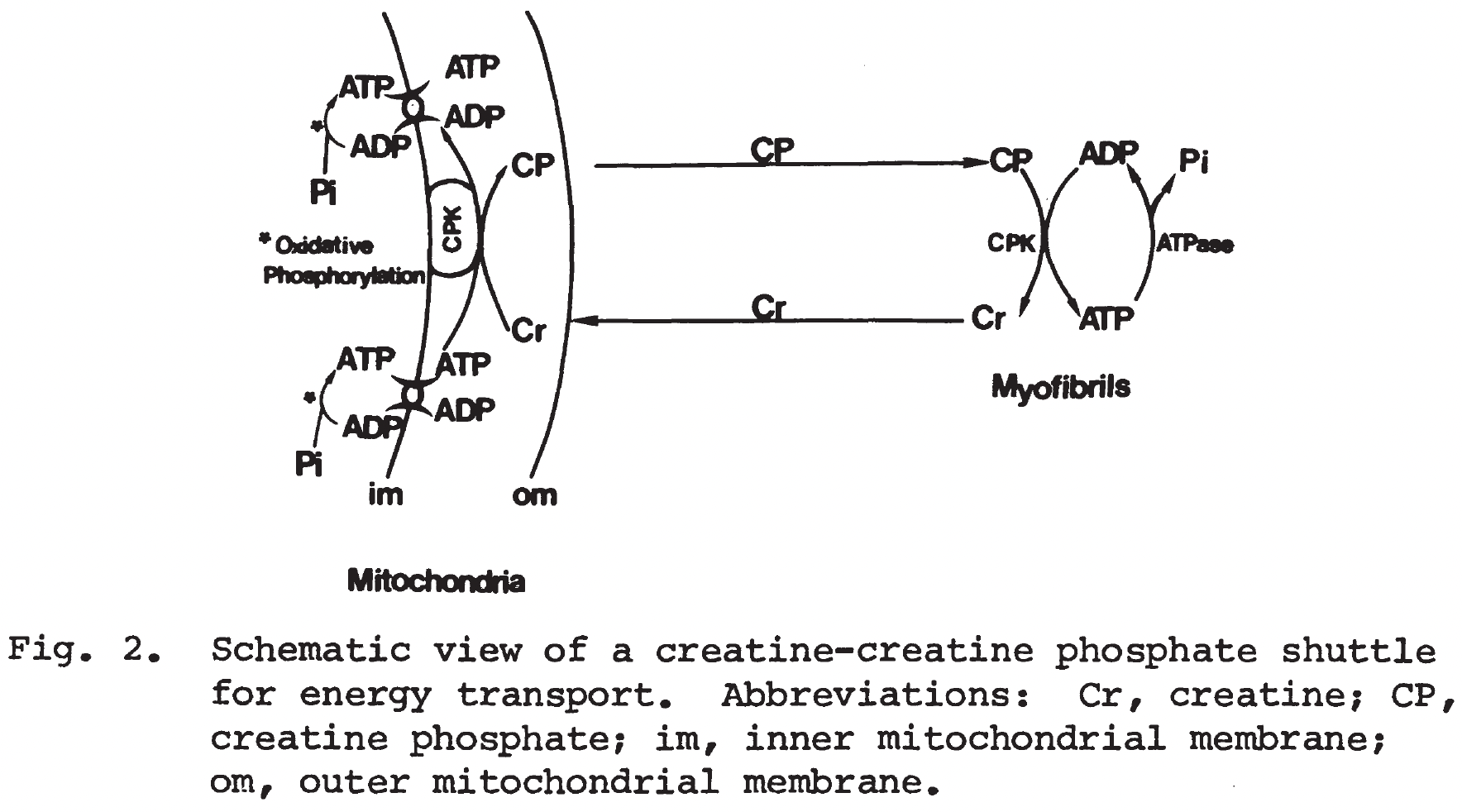
Nice work Hao. So, maybe the reaction should be changed, is that what you are proposing? I'm still a bit confused by this. It seems there are many isozymes, some which operate in the cytosol (then generating ATP) and some that operate "in" mitochondria that consume ATP. Right now, I feel like I don't dare to change the reaction since things are still a bit unclear.
To implement the schematic view, technically the reactions MAR08427, MAR08428 and MAR08430 should be modified by changing metabolites involved in [m] to [i], while the GPR of MAR08427 probably should also be adjusted to the corresponding isozymes.
These changes shouldn't affect model's performance, and would fit the commit did in this PR.
To my understanding it is costly to move ATP out of mitochondria (with ATP translocase), and I suspect this somehow also holds if you go via creatine phosphate - we need to check that we don't mess that up.
@johan-gson look forward to your test results
This will unfortunately have to wait, I only have 8 days left at SysBio and a lot of things to do :)
Sure, all the best!
@haowang-bioinfo I wonder if we shouldn't just cancel this, both the PR and the issue? My original idea was just not right, and I will not find any energy to dig into this anytime soon.
@johan-gson @haowang-bioinfo we can close for now and always re-open later if necessary.
sure, let's close it