fastp
 fastp copied to clipboard
fastp copied to clipboard
Duplication rate differs up to 30% from that of Fastqc for single end reads
I found a quite big difference in the duplication between Fastp and Fastqc. For all my ~40 SE RNAseq samples, the rate is around 10-30% lower in Fastp compared to Fastqc. Is there an explanation for this or is it a bug?

1, duplication evaluation for SE data is always inaccurate, since you never know where is the another end of the DNA fragment. For RNA data, it is more serious.
2, I found FastQC used trimmed data, the trimming will increase duplication level since it make it shorter. You can compare the original FASTQ.
@sfchen
Fastqc (Trimmed) refers to the output of Fastp which I ran only with default parameters. Sorry for the confusion.
Has anyone else seen the same discrepancy between Fastqc and Fastp?
But fastp uses the original data to calculate duplication level.
@sfchen
Multiqc says that the Fastp duplication rate was calculated on the "filtered" reads which I understand as the reads outputted by Fastp. Am I understanding something wrong or is this misleading description in Multiqc?

I also checked the Fastqc duplication rate in the original (raw) data and it corresponds almost exactly with the Fastqc duplication rate after Fastp processing.
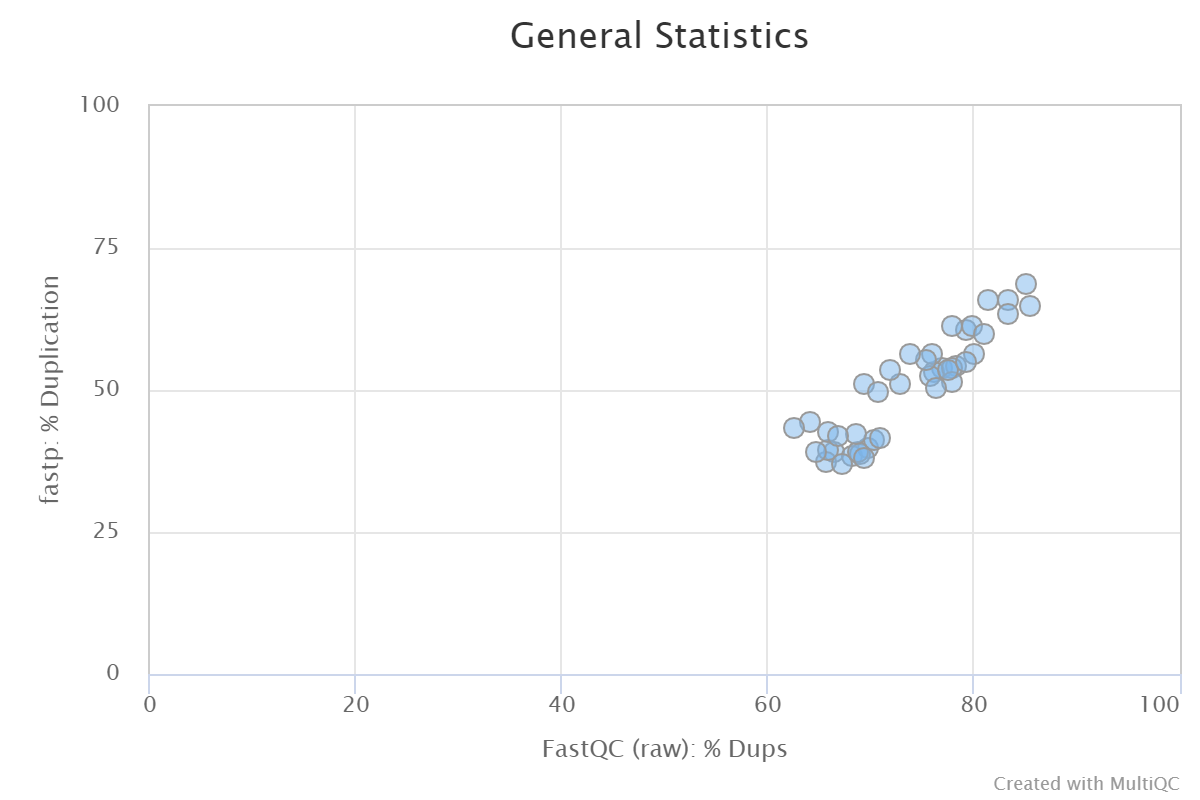
Fastp duplication rate was calculated on the original data.
I think you missed this: I also checked the Fastqc duplication rate in the original (raw) data and it corresponds almost exactly with the Fastqc duplication rate after Fastp processing.
Hi, I am using this issue as I am facing a similar problem. For SE reads the duplication rates reported by fastp and FastQC (on the same raw reads) differ significantly:
fastp --in1 test.fq.gz --out1 test_fastp.fq.gz
Detecting adapter sequence for read1...
No adapter detected for read1
Read1 before filtering:
total reads: 20271682
total bases: 3040752300
Q20 bases: 3040752300(100%)
Q30 bases: 3040752300(100%)
Read1 after filtering:
total reads: 20271682
total bases: 3040752300
Q20 bases: 3040752300(100%)
Q30 bases: 3040752300(100%)
Filtering result:
reads passed filter: 20271682
reads failed due to low quality: 0
reads failed due to too many N: 0
reads failed due to too short: 0
reads with adapter trimmed: 0
bases trimmed due to adapters: 0
Duplication rate (may be overestimated since this is SE data): 3.83467%
JSON report: fastp.json
HTML report: fastp.html
fastp --in1 test.fq.gz --out1 test_fastp.fq.gz
fastp v0.23.1, time used: 114 seconds
Running FastQC on the same input file I get the following result:
fastqc test.fq.gz -o qc/
Any ideas?
maybe try this computation mentioned here: https://www.biostars.org/p/83842/
