meanImg, meanImgE source error in skipping registration
Describe the issue:
Hi,
I want to run s2p on a single plane 2P dataset that has already been registered. The registration was done with s2p in a previous run so there is a data.bin file that contains the registered data. I wanted to use data.bin rather than registered tif to same time used on tif-to-bin conversion.
So I created a folder and moved the data.bin inside, and set ops['save_path0'] to this folder. For this s2p run, I created a new ops.npy with ops['do_registration'] = False. As expected, s2p will not run unless I specify a valid ops['data_path']. So I provided a path to tif files. However, I accidently gave the wrong data path, which lead to some ineteresting observations (see context for issue) and a question:
Question: when skipping regsitration, what does s2p use to compute ops['meanImg'] and ops['meanImgE']? Is it meant to be calculated from data_raw.bin or data.bin?
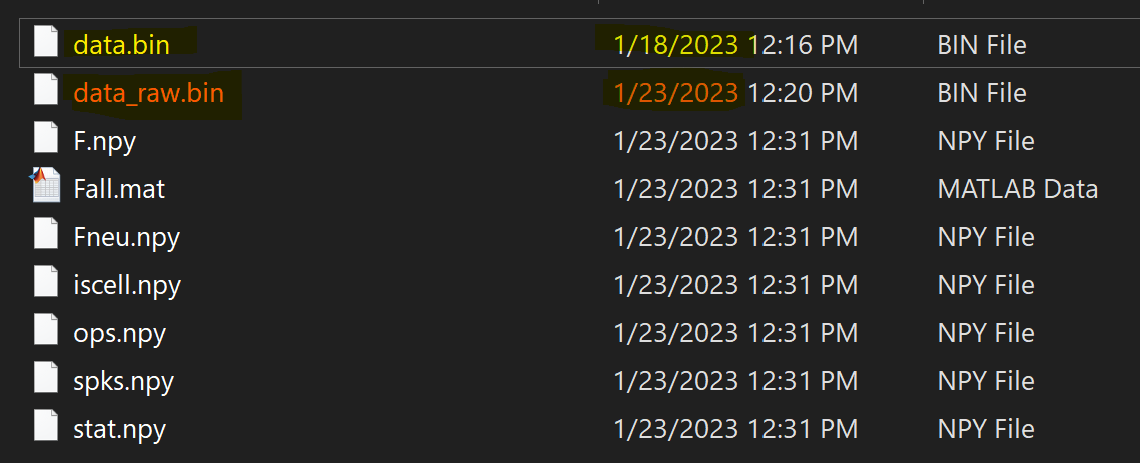
Observations: In the screenshot of the suite2p output folder:
data_raw.binis created upon this s2p run, from files inops['data_path'], which is a wrong path and contains a different dataset.data.binhas a different creation date. It is thedata.binthat contains the registered data which I copied from a previous s2p run. Sincedata.binwas found andops['do_registration'] = False, this file was not overwritten. All makes sense.- in summary,
data_raw.binanddata.bincontains different datasets.
s2p output folder screenshot
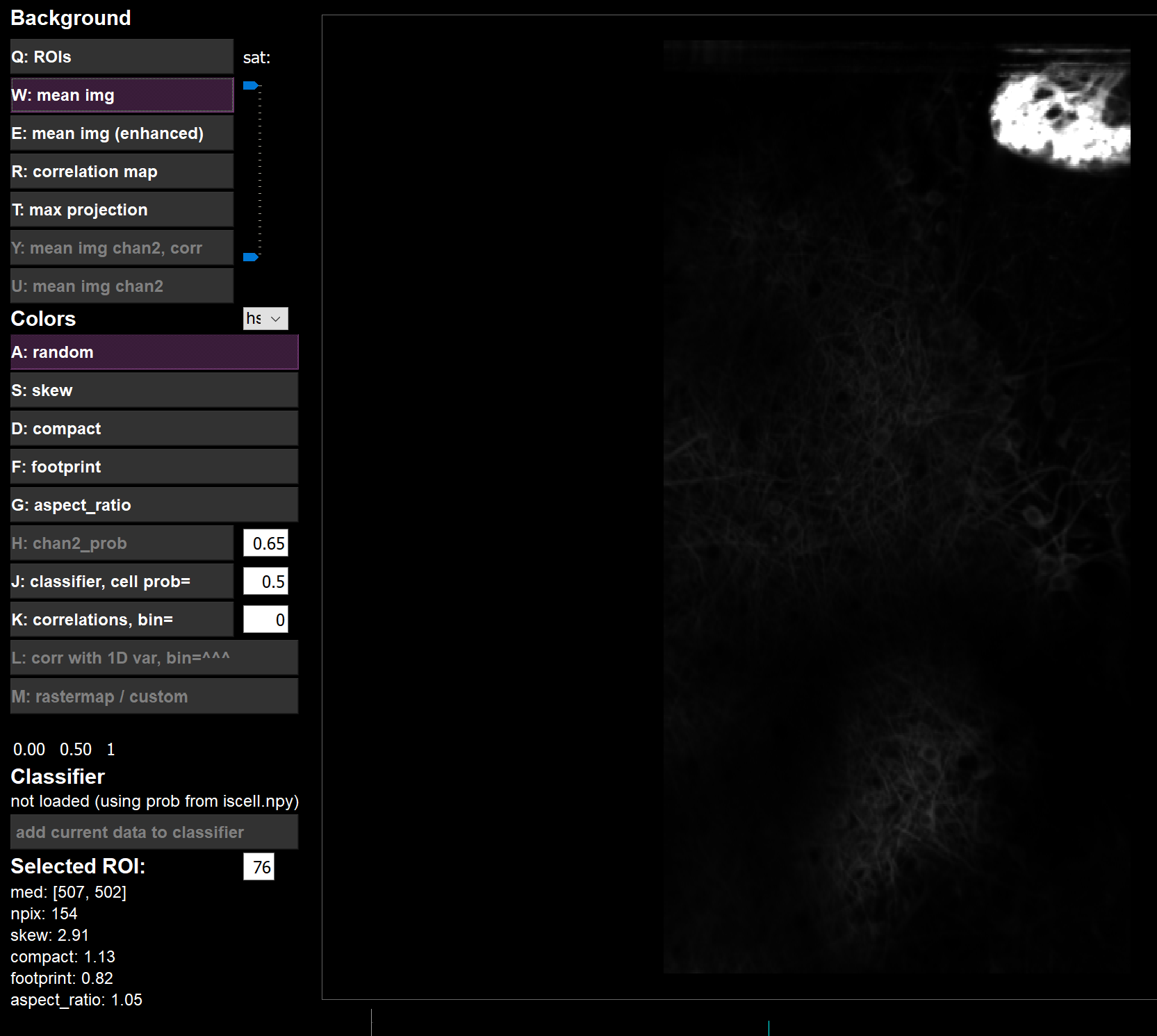
I opened the stats.npy in GUI:
- I was expecting
meanImg,meanImgE,Vcorrandmax_projto be calculated romdata.bin, anddata_raw.binwill simply be ignored, because on s2p's docmentation page, it states:
meanImg: mean of **registered** frames
meanImgE: a median-filtered version of the mean image
Vcorr: correlation map (computed during cell detection)
- Instead,
meanImg,meanImgEseem to be calculated fromdata_raw.bin
meanImg:
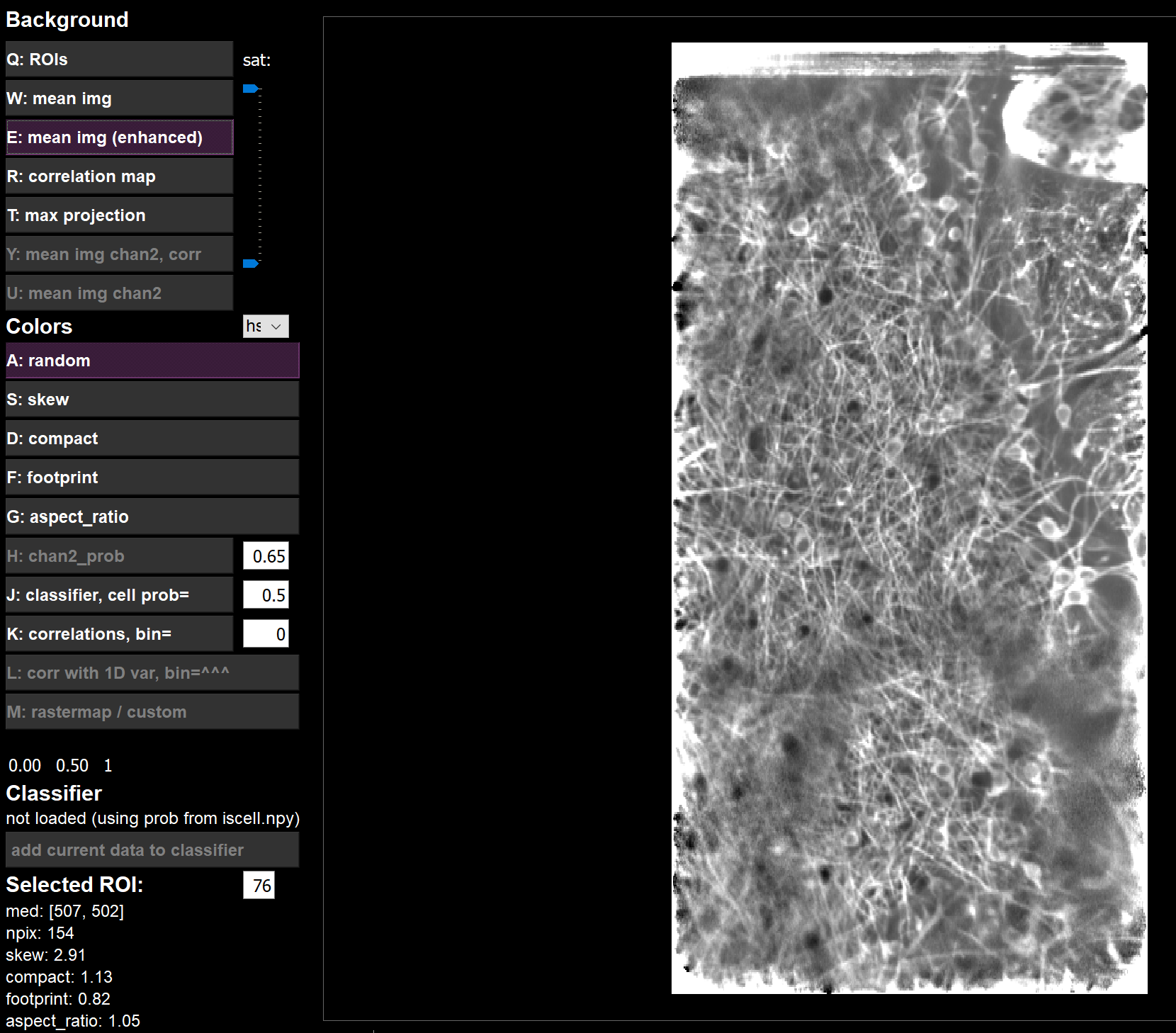
meanImgE
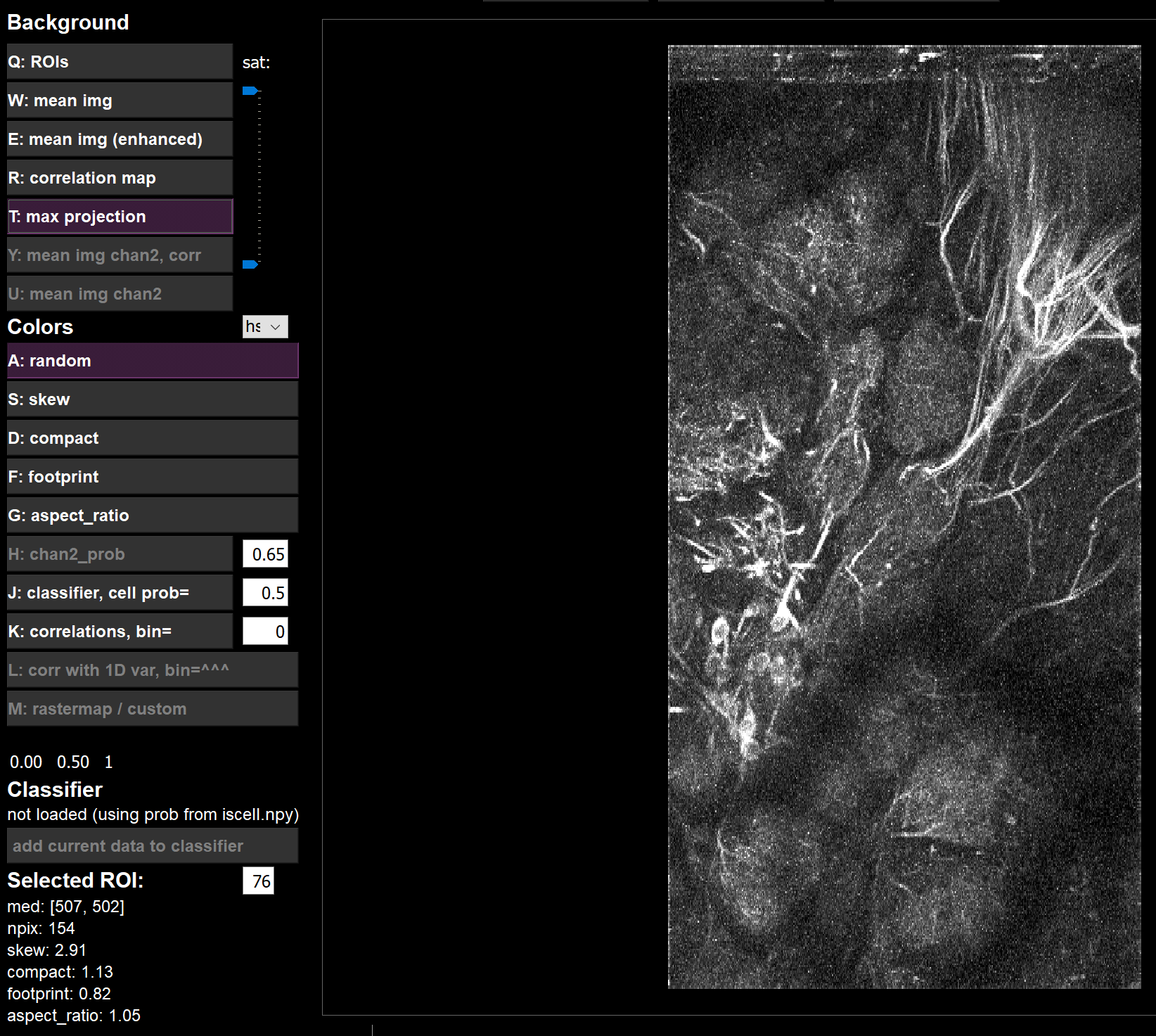
Vcorrandmax_projare fromdata.binas expected
Vcorr

max_proj
To re-iterate my question: if I skip registration in s2p, is data_raw.bin involved in any computation?
If I understand s2p correctly, all computations after registration will be using the registered datasets stored in data.bin. Then the above is simply a bug associated with skipping registration...
Thank you very much!
Reproduce the code example:
# ops definition
ops['suite2p_version'] = '0.11.1'
ops['save_path0'] = '/camp/project/Y391/0-s2p/7-detect_TC/test_detectTC_02'
ops['nplanes'] = 1
ops['nchannels'] = 1
ops['functional_chan'] = 1
ops['tau'] = 0.7 # for GCaMP6f
ops['fs'] = 2.50 # as shown on scanImage, was 2.576 for sciScan, why differ?
ops['frames_include'] = -1 # specifes frames to process, useful to test param on subset of dat, -1 means all
ops['multiplane_parallel'] = False # see debug/error 2
ops['ignore_flyback'] = [-1] # a list, 0-indexing of plane to ignore, -1 means process all
ops['preclassify'] = 0 # only extract signals for rois passing thr, 0 means all ROIs kept and signals
ops['save_mat'] = True
ops['combined'] = True
ops['aspect'] = 0.5 # 1 for 2P.v.2, use 0.5 for 2P.v.1
ops['look_one_level_down'] = False
ops['fast_disk'] = [] # location for temporary binary file
ops['delete_bin'] = False # whether or not to delete binary file created during registration
ops['mesoscan'] = False
ops['bruker'] = False
ops['bruker_bidirectional'] = False
ops['h5py'] = []
ops['h5py_key'] = 'data'
ops['nwb_file'] = ''
ops['nwb_driver'] = ''
ops['nwb_series'] = ''
ops['save_folder'] = []
ops['subfolders'] = []
ops['move_bin'] = False # use to set final bin file destination
ops['force_sktiff'] = False # whether or not to use scikit-image to read tiff
ops['save_NWB'] = False
ops['do_bidiphase'] = False # see debug/error 3
ops['bidiphase'] = 0 # in px, non-0 values will overwrite do_bidiphase
ops['bidi_corrected'] = False
ops['report_time'] = False # time dict for each plane
# REGISTRATION ---------------------
ops['do_registration'] = False
ops['keep_movie_raw'] = True # can view in registered and non-registered bin in GUI
ops['two_step_registration'] = False # for low SNR (needs keep_movie_raw = True); try without first
ops['align_by_chan'] = 1
ops['nimg_init'] = 3000 # larger may give better SNR but may blur cells
ops['batch_size'] = 700 # TA legacy, larger runs fatser but needs more RAM
ops['maxregshift'] = 0.1 # visual inspection shows considerable shifts, give high allowance
ops['smooth_sigma'] = 1.15 # in px, smoothing
ops['smooth_sigma_time'] = 1 # in frames, need 1, 2 for low SNR
ops['reg_tif'] = True # whether or not to write registered bin to tif
ops['reg_tif_chan2'] = False
ops['subpixel'] = 10 # 1/10 subpixel steps, sets precision of subpixel reg
ops['th_badframes'] = 1 # thr to exclude frames for cropping
ops['norm_frames'] = True # normalise frames when detecting shifts
ops['force_refImg'] = False # whether to use given refImg
ops['refImg'] = '' # path for refImg
ops['pad_fft'] = False # whether or not to pad image during FFT portion of reg
# NON-RIGID ------------------------
ops['nonrigid'] = True # False does not work...
ops['block_size'] = [128, 128] # in px, keep as a power of 2 or 3 (128, 256, 384 etc)
ops['snr_thresh'] = 1.5 # set to high value for low SNR
ops['maxregshiftNR'] = 3 # in px, max. block shift relative to rigid shift
# 1P - irrelevant
ops['1Preg'] = False
ops['spatial_hp'] = 42
ops['spatial_hp_reg'] = 42
ops['pre_smooth'] = 0
ops['spatial_taper'] = 40
# ROI DETECTION ---------------------------------------------
ops['roidetect'] = True
ops['sparse_mode'] = False
ops['spatial_scale'] = 3 # 0 is automatic, if off, set manually 2=12px, 3=24 px
ops['connected'] = True # whether or not ROIs needs to be fully connected (use 0 for den or bouton)
ops['threshold_scaling'] = 1 # use low value to reject less ROIs
ops['spatial_hp_detect'] = 25 # ? window for spatial high-pass filtering for neuropil subtracation before ROI detection takes place
ops['max_overlap'] = 0.3
ops['high_pass'] = 100 # window of mean subtration in time
ops['smooth_masks'] = True # whether to smooth masks in final round of detection, useful for low SNR
ops['max_iterations'] = 100 # set high is TA legacy
ops['nbinned'] = 5000 # max. binned frames to use for roi detection
ops['denoise'] = False # whether or not to denoise binned movie before roi detection, need sparse_mode=True; 1 gave funky shapes in Y399
ops['chan2_thres'] = 0.65
# CELLPOSE DETECTION (not used) ----------------------------------------
# these settings only applied if anatomical_only > 0
ops['anatomical_only'] = 0 # 3 = using enhanced mean image
ops['diameter'] = [12, 24] # 0 means estimate diameter
ops['cellprob_threshold'] = 0 # ?
ops['flow_threshold'] = 1.5 # ?
ops['pretrained_model'] = 'cyto' # path to pretrained model
# SIGNCAL EXTRACTION ------------------------------------
ops['neuropil_extract'] = True
ops['allow_overlap'] = False # whether to extract trace from pixels belong to multiple ROIs
ops['inner_neuropil_radius'] = 2 # px to keep between ROI and neuropil donut
ops['min_neuropil_pixels'] = 350 # min. px used to compute neuropil for each ROI
ops['lam_percentile'] = 50 # ? Percentile of Lambda within area to ignore when excluding cell pixels for neuropil extraction
# SPIKE DECONVOLUTION - irrelevant
ops['spikedetect'] = False
ops['neucoeff'] = 0.7
ops['baseline'] = 'maximin'
ops['win_baseline'] = 60
ops['sig_baseline'] = 10
ops['prctile_baseline'] = 8
# CLASSIFICATION - irrelevant
ops['soma_crop'] = True # whether to crop dendrites for cell classification stats
ops['use_builtin_classifier'] = False
ops['classifier_path'] = ''
Error message:
# .out from s2p run
{'data_path': '/camp/project/Y391/reg_tif/plane5', 'save_path0': '/camp/project/Y391/plane2', 'tiff_list': ['/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk0_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk1_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk2_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk3_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk4_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk5_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk6_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk7_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk8_plane5.tif', '/camp/project/Y391/reg_tif/plane5/Y391_s2p_test03_full_refplane4_reg_chunk9_plane5.tif']}
tif
** Found 10 tifs - converting to binary **
2800 frames of binary, time 10.11 sec.
time 167.54 sec. Wrote 27880 frames per binary for 1 planes
>>>>>>>>>>>>>>>>>>>>> PLANE 0 <<<<<<<<<<<<<<<<<<<<<<
NOTE: not running registration, ops['do_registration']=0
binary path: /camp/project/Y391/plane2/suite2p/plane0/data.bin
NOTE: Applying builtin classifier at /camp/.conda/envs/suite2p_v0_11_1/lib/python3.8/site-packages/suite2p/classifiers/classifier.npy
----------- ROI DETECTION
Binning movie in chunks of length 05
Binned movie of size [5576,512,512] created in 35.20 sec.
ROIs: 184, cost: 0.7039, time: 235.5386
ROIs: 318, cost: 0.6833, time: 307.9646
ROIs: 366, cost: 0.6763, time: 368.2374
ROIs: 378, cost: 0.6744, time: 421.7713
ROIs: 378, cost: 0.6310, time: 481.3987
ROIs: 378, cost: 0.6167, time: 508.4664
ROIs: 378, cost: 0.6150, time: 537.3987
Detected 378 ROIs, 596.85 sec
After removing overlaps, 294 ROIs remain
----------- Total 633.20 sec.
----------- EXTRACTION
Masks created, 3.12 sec.
Extracted fluorescence from 294 ROIs in 27880 frames, 45.02 sec.
----------- Total 48.32 sec.
----------- CLASSIFICATION
['compact', 'skew', 'npix_norm']
WARNING: skipping spike detection (ops['spikedetect']=False)
Plane 0 processed in 682.31 sec (can open in GUI).
total = 849.86 sec.
TOTAL RUNTIME 849.86 sec
Version information:
suite2p v0.11.1
Context for the issue:
No response