How to use the command line to get the same information (cell number) displayed in the GUI
First of all, thank you very much for maintaining cellpose, it helps me to get things done quickly.This is a great job.
In my work I ran into a problem: how to get the same information (cell number) that I get after batch processing the pictures using the command line and get a single photo after segmenting it in the GUI.
Although the segmentation effect I get with the command line is the same as the GUI version, I have tried many methods with imageJ and can't get the same number of cells as the GUI version shows.
Here is what shows in the GUI.
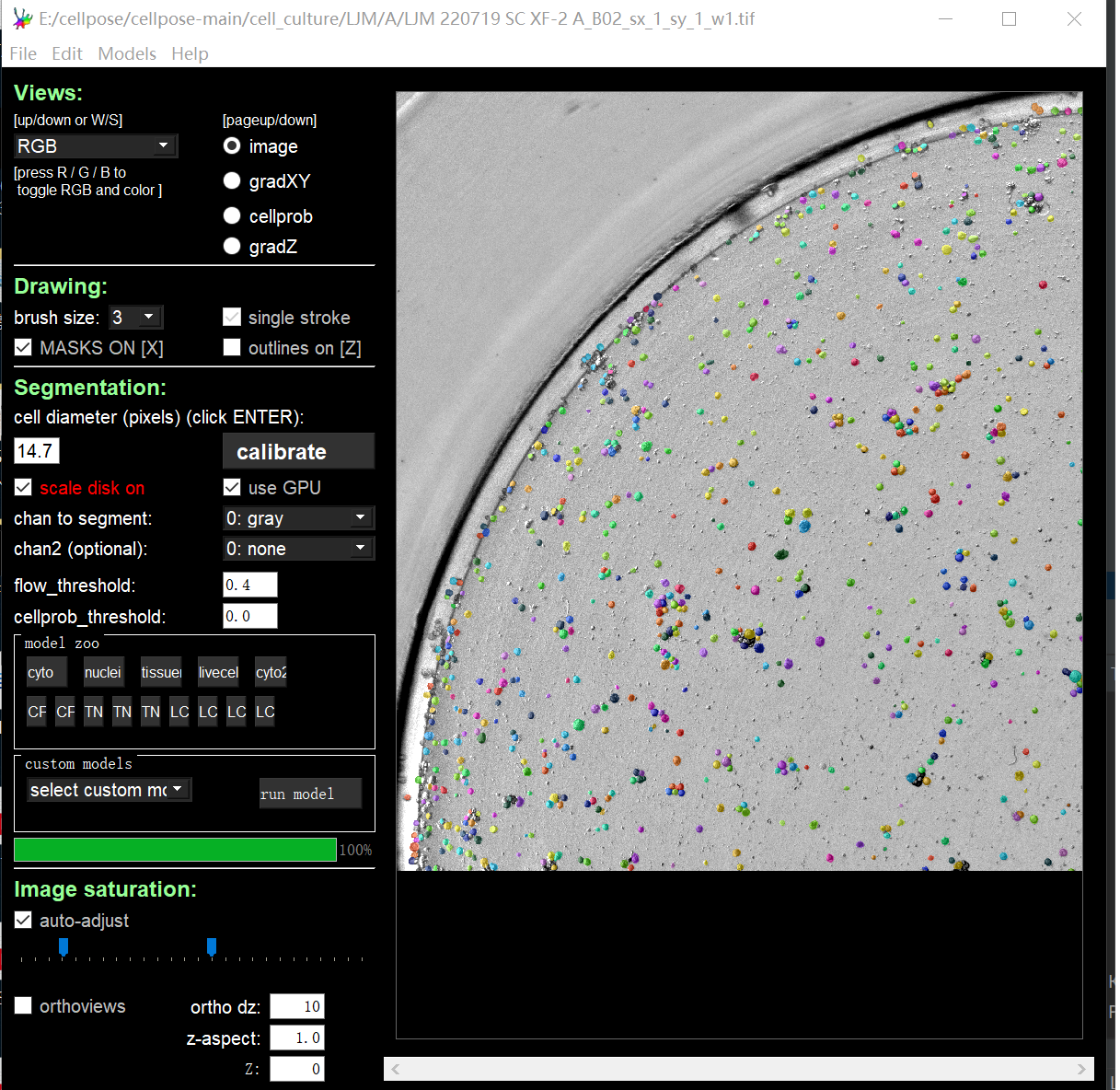
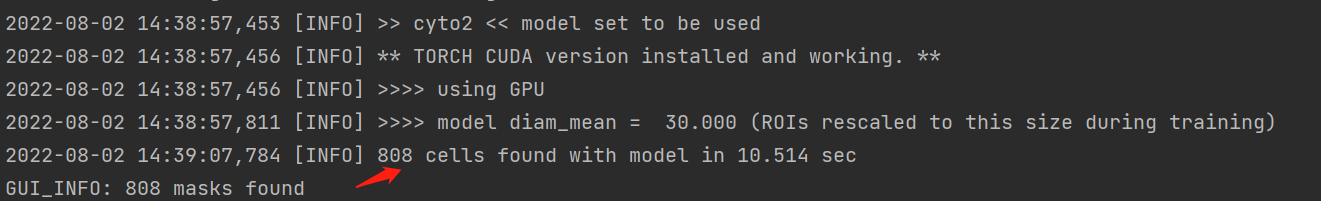
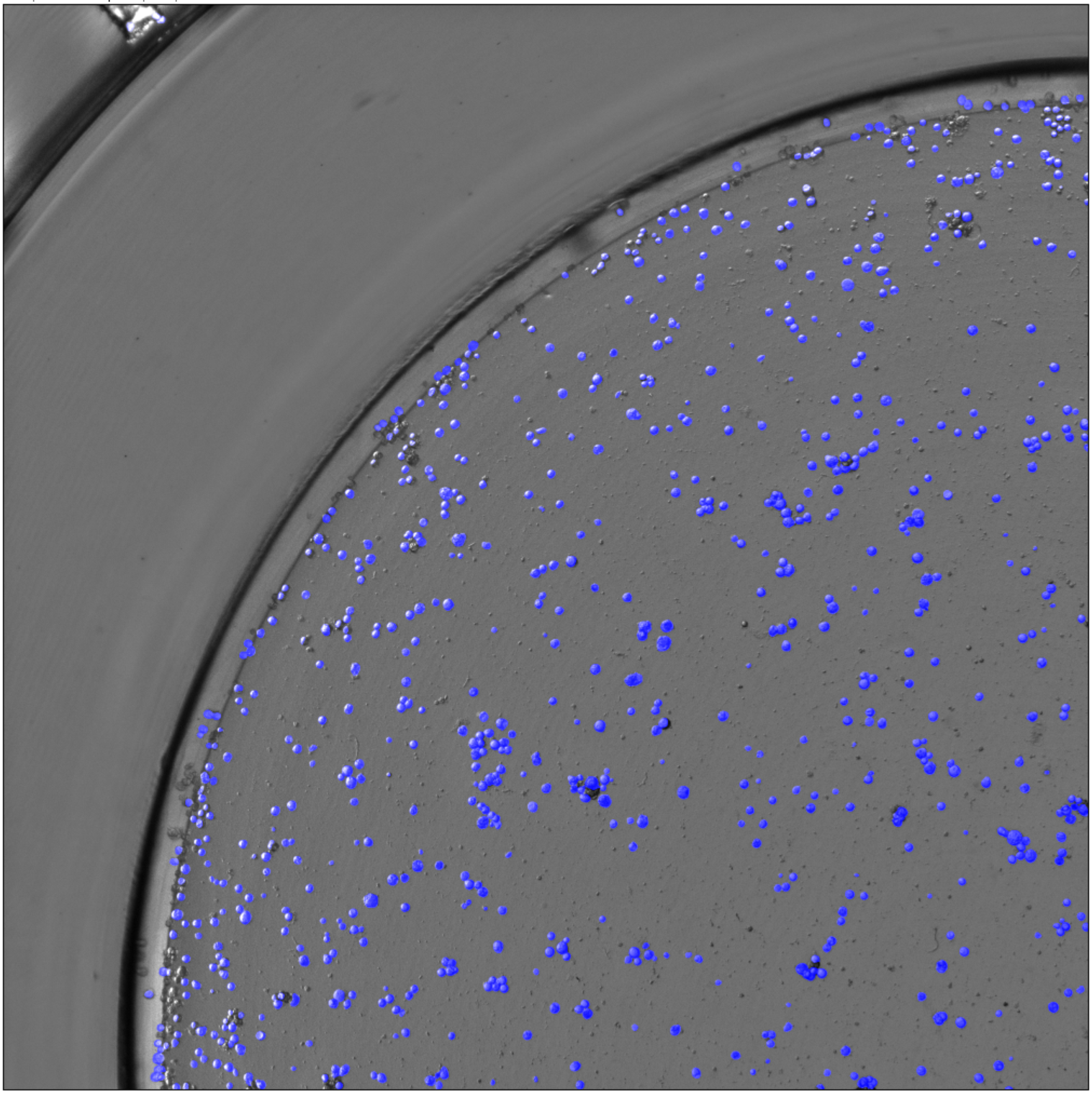
This is the mask picture I got with the command line when batch processing, and the count result I got from imageJ (I have used many segmentation methods and can't match the GUI display).
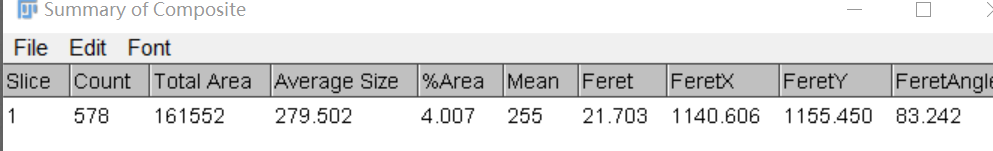
Or is there any way to print the cell count information when batch processing images ?
hmm this is a very common request. what if there is an option to make a txt file in the --dir that outputs two columns: the name of each file and the number of ROIs?
regarding imageJ import, are you using the --save_outlines option?
I'd also be interested in this feature, were it to be added. We're hoping to export a list of ROIs from images directly from the Colab notebook. Currently, we're having to save to '_seg.npy', download, and load into the GUI to view the number of cells.
Resolved by adding:
for mask in masks:
print(len(utils.outlines_list(mask)))
Sounds like this is related to #581? @carsen-stringer, let me know what you need to complete #643 and get this functionality added.
thanks @mrariden , closing this issue as it will be resolved with #643