usher
 usher copied to clipboard
usher copied to clipboard
Web Error: "Why did we not get EOF starting out with bignum"
@shay671 mentioned to me that he gets a weird error using USHER in the browser
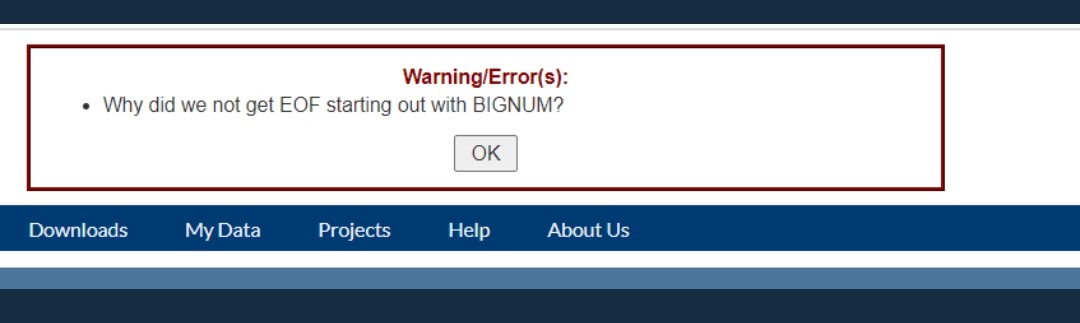
Also encountered by @ryhisner I think
Thanks -- so sorry about that, at >12 million sequences, the protobuf byte size has exceeded a constant that seemed safe enough when I wrote the code 2 years ago! It is fixed on our test server (https://genome-test.gi.ucsc.edu/cgi-bin/hgPhyloPlace) and should be fixed on the main site (https://genome.ucsc.edu/cgi-bin/hgPhyloPlace) very soon.
Also found by @agamedilab and probably @FedeGueli too.
Wow, Indeed, who had thought two years ago that we would exceed 12M samples of SARS-CoV-2 sequences?
Thank you, @AngieHinrichs, for your amazing work. It is a breakthrough that enables so many independent researchers to investigate and learn.
It is fixed on the main site https://genome.ucsc.edu/cgi-bin/hgPhyloPlace
Thanks again for reporting the error! For future reference, bugs in the web interface can also be reported to the UCSC Genome Browser list [email protected] .
And thanks @shay671! I couldn't do it without @yatisht's, @yceh's and @jmcbroome's work on UShER, matOptimize and matUtils, and we're so happy that people are finding it useful.