usher
 usher copied to clipboard
usher copied to clipboard
BUG: Multiple mutations in one codon cause can cause false amino acid mutation
When looking at sequence Iran/AN127/2022|EPI_ISL_10195257, I noticed that it was falsely annotated with Q498*.
Nextclade annotates it with Q498Q:
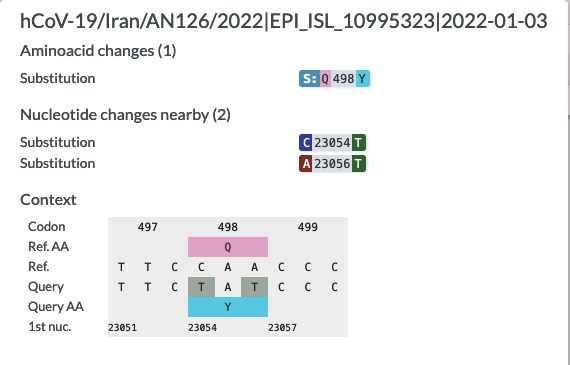
The reason for Usher's false annotation seems to be the presence of two mutations in one codon.
On an internal branch, I could even see two mutations listed side by side Q498H, Q498* which is clearly not possible.
How do you deal with multiple mutations in one codon? Is this a known limitation by design?
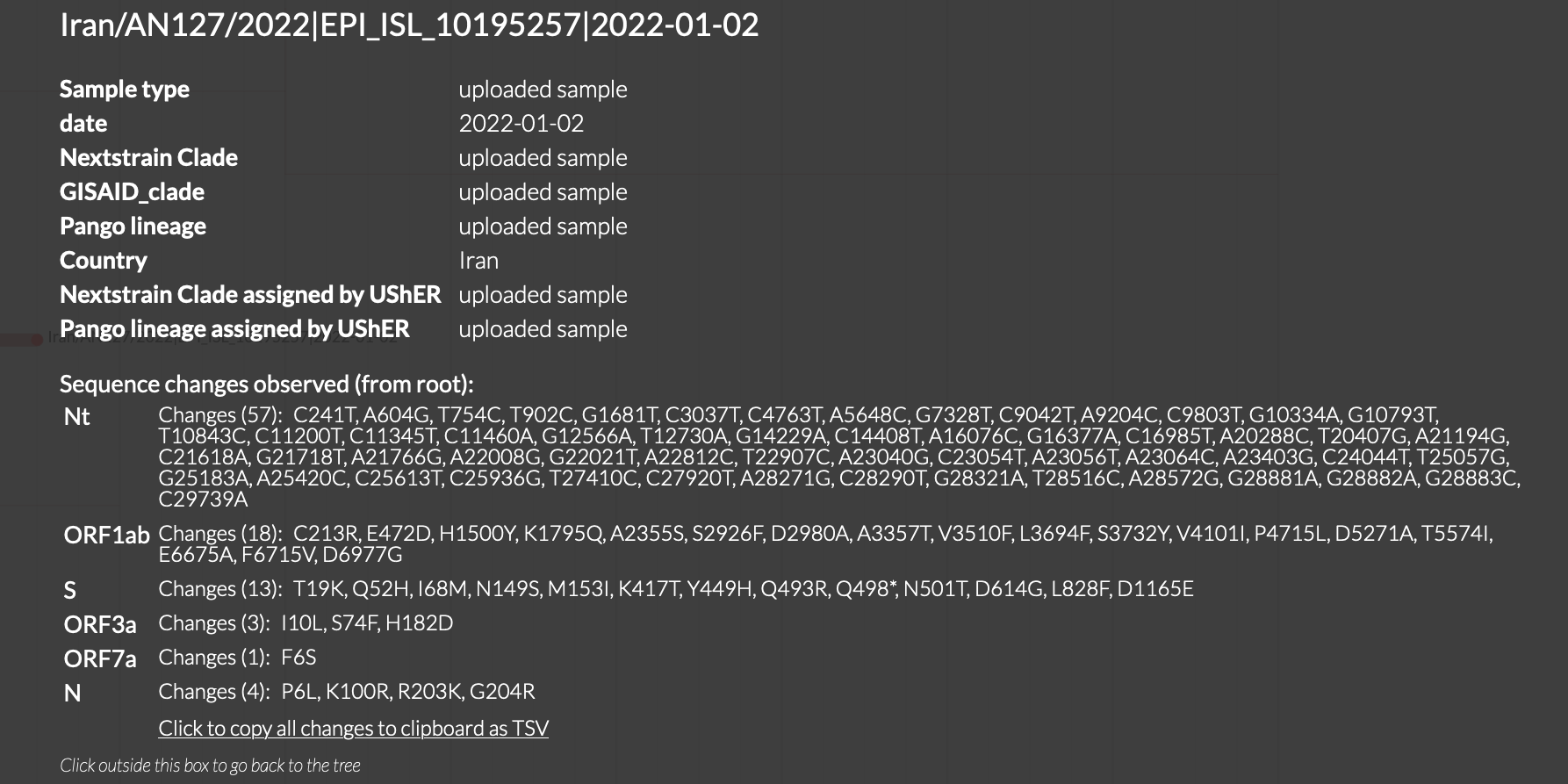
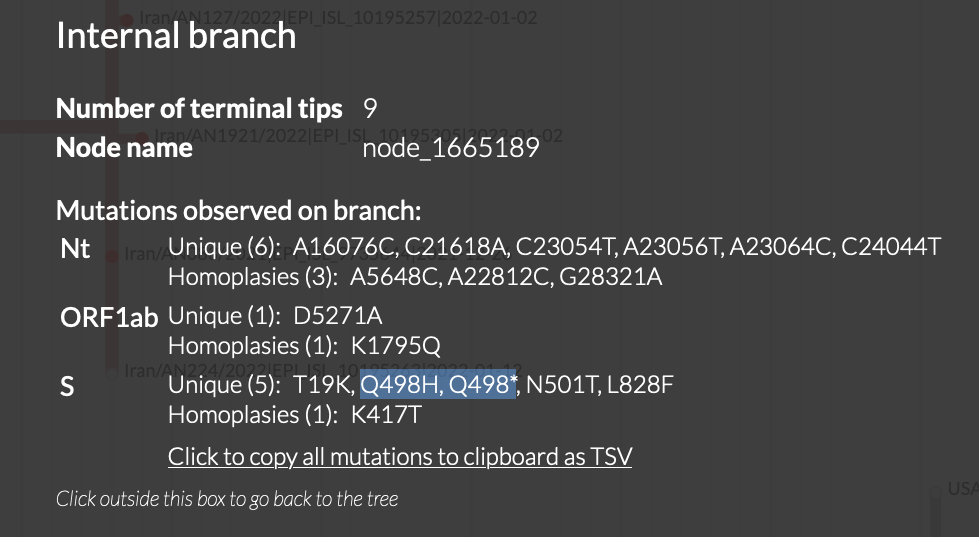
Sorry, the web interface uses simple code that translates each nucleotide substitution into its corresponding amino acid substitution independently. I'll add it to the list of badly needed improvements but am not sure when I'll have time to work on it.
I believe this is fixed on dev.usher.bio now - CV.2 and XBB.1.5 are good for testing this. Also a cluster in XBC.2 that has a (probably false) reversion on 23019 that causes S:P486L (e.g. Brunei/7722014505/2022|EPI_ISL_15696712|2022-10-10), with one member of that cluster regaining T23019C restoring 486P (Brunei/7722017695/2022|EPI_ISL_16231620|2022-11-28).
Exciting news, thanks so much @AngieHinrichs! I'll watch out for possible regressions. Can you link to the PR that changed the behaviour? Curious how big it was, what functions were touched, etc - to be able to test better.
Looks to be working to me. Closing for now.