usher
 usher copied to clipboard
usher copied to clipboard
Issue with a Delta sub lineage
I try to run a build on these 134 samples : list for usher issue.csv
This is the tree i got https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_23199_c07fa0.json?c=pango_lineage_usher
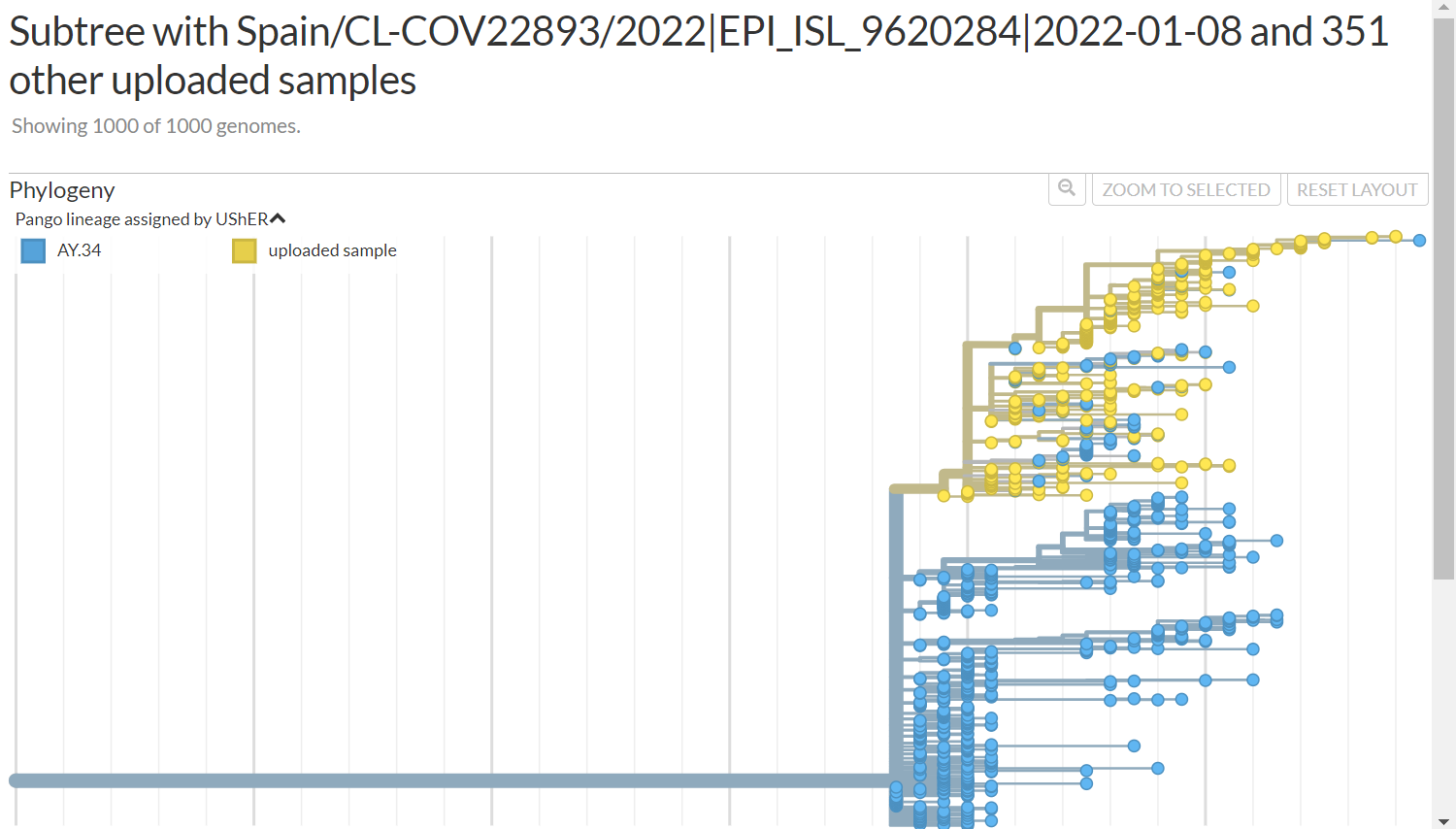
Indeed the ay.34 mutations are present in those sequence.
But, those 134 sequences share also another 8 mutations in common. When filtering them it shows different pattern on the tree :
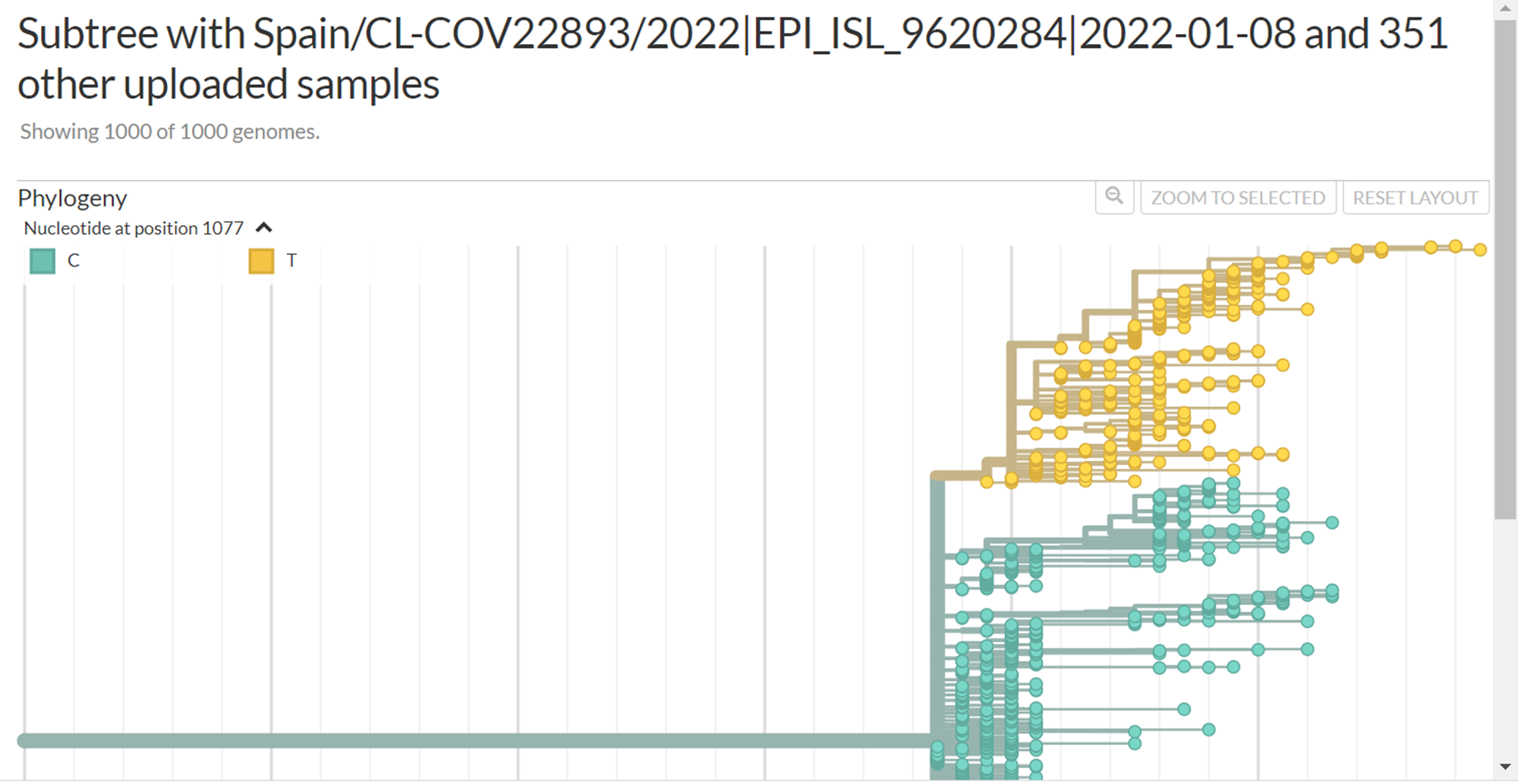
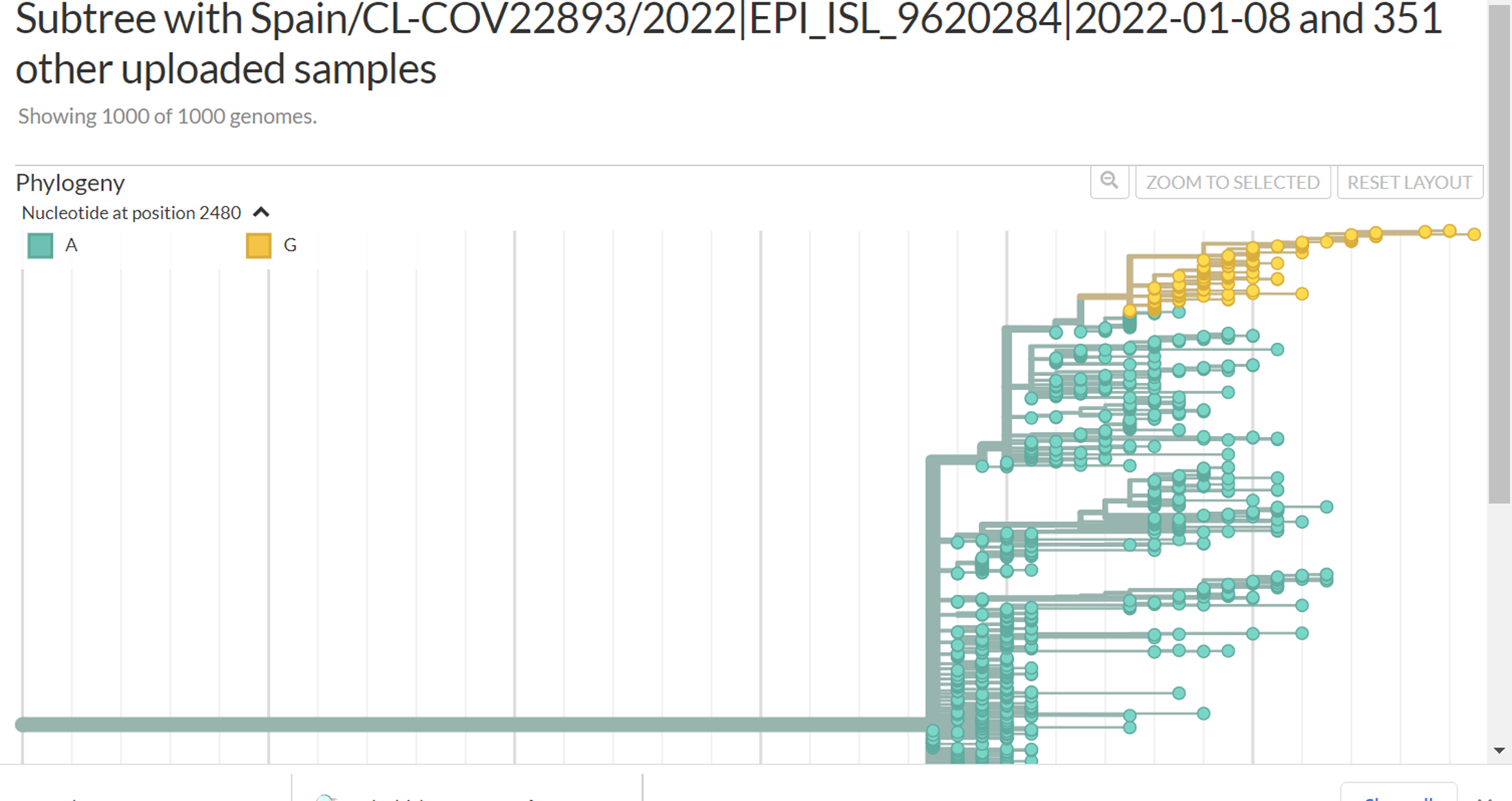
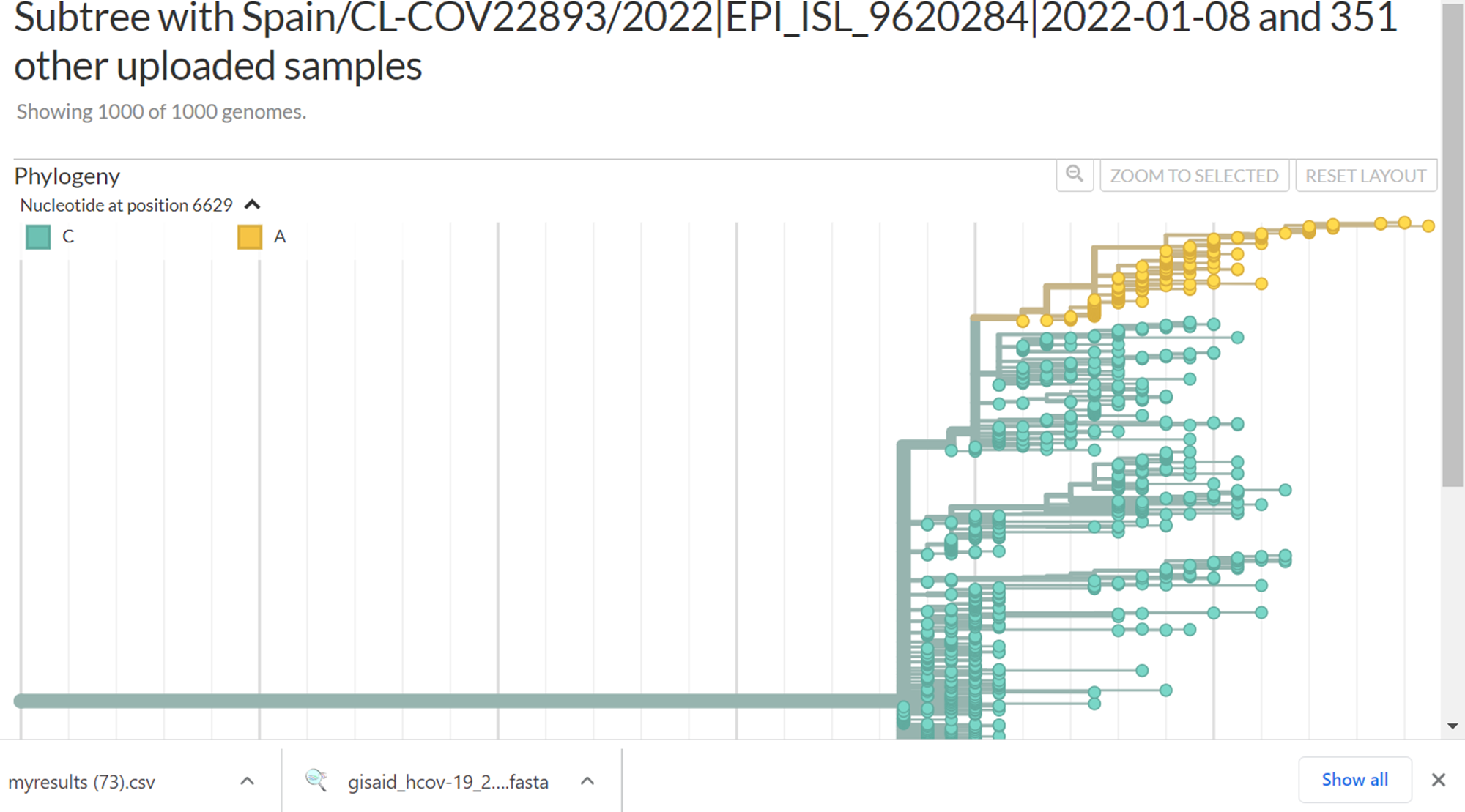
I would have expect them to cluster together with a branch of the shared mutations.
Thanks.
Hi @shay671 -- the 134 sequences in list for usher issue.csv seem to correspond only to the smaller branch with A2480G, not all uploaded samples in https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_23199_c07fa0.json?c=pango_lineage_usher . The title of the nextstrain view is "Subtree with Spain/CL-COV22893/2022|EPI_ISL_9620284|2022-01-08 and 351 other uploaded samples" -- implying that 352 samples were uploaded, not just the 134.
If I click on the branch with all uploaded sequences, click on "DOWNLOAD DATA" (near the bottom of the page), and then click the "METADATA (TSV)" option, then a file with a very long name is downloaded. If I grep uploaded in that file, there are 352 results. I extracted just the EPI_ISL_ IDs into this file: full.list.for.usher.txt
If I click on the smaller branch with mutations A2480G,T27351C, and again save metadata and extract EPI_ISL IDs for the rows that contain uploaded, then I get 145 IDs: the 134 IDs in list.for.usher.issue.csv plus these IDs:
EPI_ISL_8626123
EPI_ISL_8626692
EPI_ISL_8626694
EPI_ISL_8626828
EPI_ISL_8627374
EPI_ISL_8627488
EPI_ISL_8627698
EPI_ISL_8627701
EPI_ISL_8627712
EPI_ISL_8731499
EPI_ISL_8731523
To double-check, if I upload your file list.for.usher.issue.csv to the web interface then I get this subtree ("Spain/CL-COV16546/2021|EPI_ISL_8626820|2021-10-26 and 133 other uploaded samples", with only the smaller branch highlighted as uploaded):
https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_29023_c53d50.json?c=pango_lineage_usher
Hi @AngieHinrichs . My meaning is that i would have expected to get some branch leading to those as they share a unique pattern of mutations the other samples do not.
Yes, there is a branch containing those 134 sequences: the smallest branch in your 3 colorings by nucleotide position, that includes A2480G. Your file list for usher issue.csv contains 134 EPI_ISL_ IDs. Your tree view https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_23199_c07fa0.json?c=pango_lineage_usher was built with a larger set of 352 IDs, and some of those 352 sequences do not have all 8 mutations that are found in the set of 134. When you color by position 1077, it shows the larger branch with all 352 sequences; when you color by 2480, now you're narrowing it down to the smaller branch that contains the 134 sequences. When I uploaded your file, I got a tree in which the uploaded samples were all on the smaller branch (with a longer series of mutations leading to it than the larger branches).
Here is a view of the result of uploading only the 134 IDs, with nucleotide changes annotated on all branches:
https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/subtreeAuspice1_genome_29023_c53d50.json?branchLabel=nuc%20mutations&c=pango_lineage_usher&label=nuc%20mutations:C1077T,G5230T
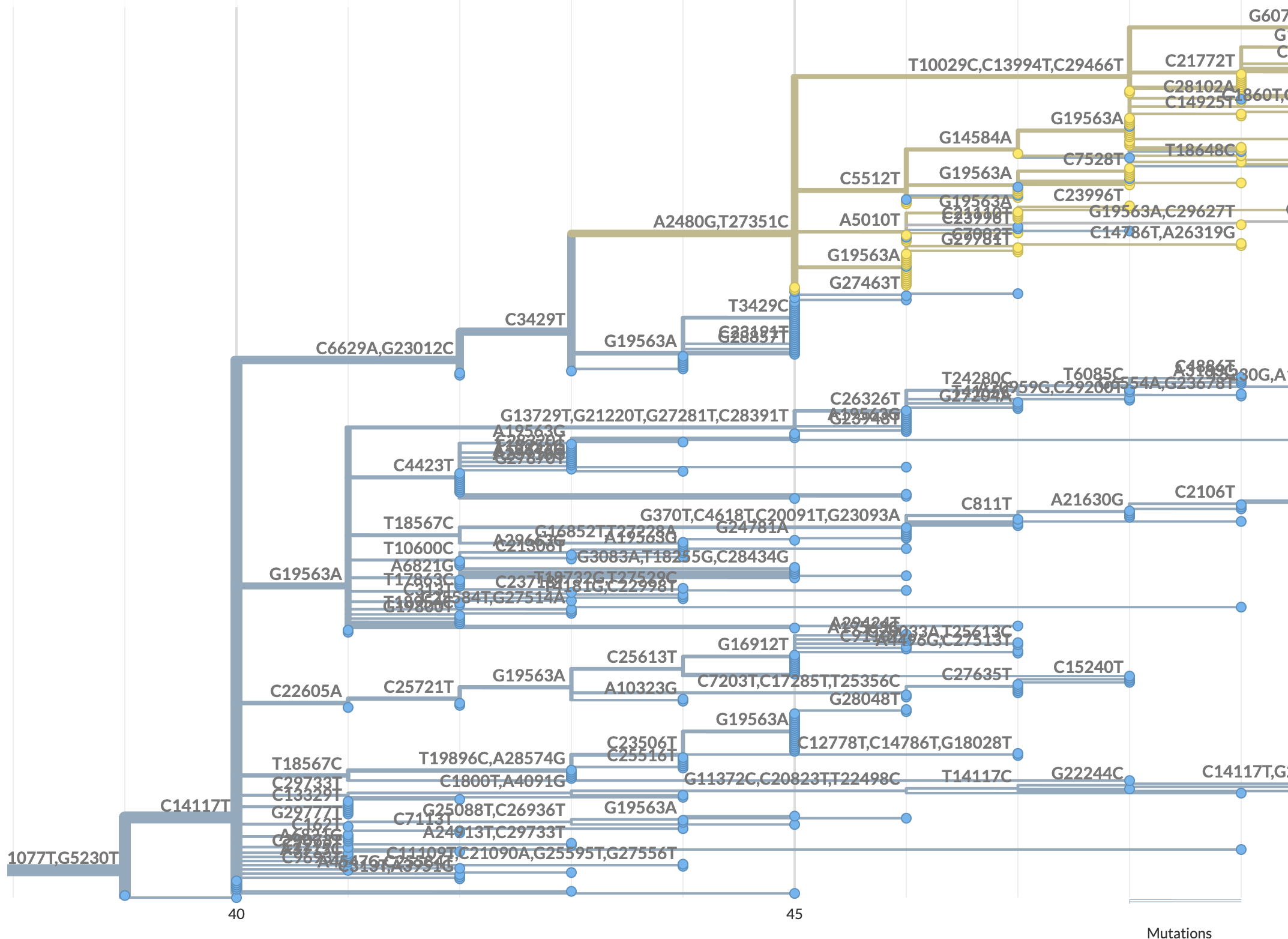
Can you send an example of a sequence name or ID from the subtree view that is not on a branch with all of the expected mutations?
Hi @AngieHinrichs I see what you mean now. You are totally correct. Thank you for your patience, that's been very helpful.