SVision
 SVision copied to clipboard
SVision copied to clipboard
SVision runs too slow...
Hello! I use SVision to run my cancer data from Pacbio long read seqencing(CLR) on PC with 28 threads, but I foung it's too slow. It takes me 3 days with 20 threads but still not finish and nothing in predict_results directory. My cancer data is about 30* coverage and mapping reference using pbmm2. And my reference is already remove other contigs, only chr1-chr22 and chrX, chrY. My code is /mnt/f/zer/software/svision/SVision-1.3.8/SVision -o /mnt/f/zer/hg38_chr1_22xy/1-complexSV/1-SVision_result/** -b /mnt/f/zer/hg38_chr1_22xy/pbmm2//_hg38_nor.bam -m /mnt/f/zer/software/svision/SVision-1.3.8/cnn_model/svision-cnn-model.ckpt -g /mnt/f/zer/reference/hg38/hg38_chr1_22xy.p13.fa -n ** -s 5 -t 20 --graph --qname Python version is 3.6.13 and my CPU utilization is about 15%, memory utilization is about 44% (112/256G). I want to know that is any error ? And what should I do to increase computational speed?
Hi,
What are the outputs in the log file, could please paste the last few lines. Moreover, please try without --graphoption and see what's going on.
It seems that no error in the log. And I try to run without --graph option as you suggested but find it did not increase computational speed more. My CPU utilization is about 24%, memory utilization is about 23% (58/256G). In top, I see there are 29 tasks total, but only 3 or 5 tasks are running, others are sleeping. with --graph option 2022-10-30 02:20:42,656 [INFO ] Processing chr9:90000000-100000000, 8444 segments write to: chr9.segments.9.bed 2022-10-30 02:29:38,718 [INFO ] Processing chr9:100000000-110000000, 7644 segments write to: chr9.segments.10.bed 2022-10-30 02:38:00,617 [INFO ] Processing chr9:110000000-120000000, 7900 segments write to: chr9.segments.11.bed 2022-10-30 02:57:44,374 [INFO ] Processing chr9:120000000-130000000, 8899 segments write to: chr9.segments.12.bed 2022-10-30 02:59:33,977 [INFO ] Processing chr9:130000000-138394717, 9580 segments write to: chr9.segments.13.bed 2022-10-30 03:12:29,141 [INFO ] Processing chrX:0-10000000, 2003 segments write to: chrX.segments.0.bed 2022-10-30 03:15:16,413 [INFO ] Processing chrX:10000000-20000000, 2745 segments write to: chrX.segments.1.bed 2022-10-30 03:47:49,344 [INFO ] Processing chrX:20000000-30000000, 2484 segments write to: chrX.segments.2.bed
without --graph option **************** Step1 Image coding and segmentation ****************** 2022-11-01 21:25:47,635 [INFO ] Processing chr1:130000000-140000000, 0 segments write to: chr1.segments.13.bed 2022-11-01 21:28:16,583 [INFO ] Processing chr1:100000000-110000000, 4987 segments write to: chr1.segments.10.bed 2022-11-01 21:28:19,854 [INFO ] Processing chr1:80000000-90000000, 4858 segments write to: chr1.segments.8.bed 2022-11-01 21:28:21,801 [INFO ] Processing chr1:70000000-80000000, 4940 segments write to: chr1.segments.7.bed 2022-11-01 21:28:31,269 [INFO ] Processing chr1:90000000-100000000, 5367 segments write to: chr1.segments.9.bed 2022-11-01 21:28:42,731 [INFO ] Processing chr1:110000000-120000000, 5691 segments write to: chr1.segments.11.bed 2022-11-01 21:30:30,349 [INFO ] Processing chr1:60000000-70000000, 8132 segments write to: chr1.segments.6.bed 2022-11-01 21:31:15,614 [INFO ] Processing chr1:50000000-60000000, 9545 segments write to: chr1.segments.5.bed 2022-11-01 21:31:21,408 [INFO ] Processing chr1:30000000-40000000, 9884 segments write to: chr1.segments.3.bed 2022-11-01 21:31:26,554 [INFO ] Processing chr1:10000000-20000000, 10124 segments write to: chr1.segments.1.bed 2022-11-01 21:31:32,321 [INFO ] Processing chr1:120000000-130000000, 15335 segments write to: chr1.segments.12.bed 2022-11-01 21:31:39,977 [INFO ] Processing chr1:20000000-30000000, 10656 segments write to: chr1.segments.2.bed 2022-11-01 21:31:46,929 [INFO ] Processing chr1:40000000-50000000, 10380 segments write to: chr1.segments.4.bed 2022-11-01 21:31:59,862 [INFO ] Processing chr1:190000000-200000000, 10079 segments write to: chr1.segments.19.bed 2022-11-01 21:32:18,714 [INFO ] Processing chr1:170000000-180000000, 10316 segments write to: chr1.segments.17.bed 2022-11-01 21:32:46,780 [INFO ] Processing chr1:160000000-170000000, 10813 segments write to: chr1.segments.16.bed 2022-11-01 21:32:52,958 [INFO ] Processing chr1:180000000-190000000, 11089 segments write to: chr1.segments.18.bed 2022-11-01 21:32:54,409 [INFO ] Processing chr1:200000000-210000000, 11170 segments write to: chr1.segments.20.bed 2022-11-01 21:33:08,191 [INFO ] Processing chr1:150000000-160000000, 11607 segments write to: chr1.segments.15.bed 2022-11-01 21:33:16,341 [INFO ] Processing chr1:0-10000000, 14145 segments write to: chr1.segments.0.bed 2022-11-01 22:46:22,557 [INFO ] Processing chr1:140000000-150000000, 75033 segments write to: chr1.segments.14.bed
The number of segments identified in the 10mb window was much more than the number identified in our data. Since SVision codes these identified segments as images, the unexpected large number of segments makes the process very slow.
But it is so strange that SVision identifies such great number of segments. Here are some numbers in our analysis based on minimap2 and ngmlr alignments:
- For the ~25X HiFi data of cell lines sequenced by HGSVC, SVision only identified around 1000 segments in the chr1:200000000-210000000, where 11170 segments were identified from your data.
- For the ~60X HiFi data of tumor tissue, SVision identified around 2358 segments in the chr1:200000000-210000000.
CLR might be an issue, but we also tried ~30X ONT data and the number of identified segments look normal. I am very curious about what kind of samples you are processing. Moreover, would you please looked at the segments in the chr1.segments.20.bed, and you could go to some loci where segments are identified. It would be great if you could make screenshots of some loci in IGV.
In my cancer data, I call about 20000 SVs in per sample and about 10-20% is translocation, the proportion of translocation is higer than other cancers, it will be an issue?
In the file chr1.segments.20.bed, I replace "+" with "\t" and import it to IGV because IGV can not know the raw file.
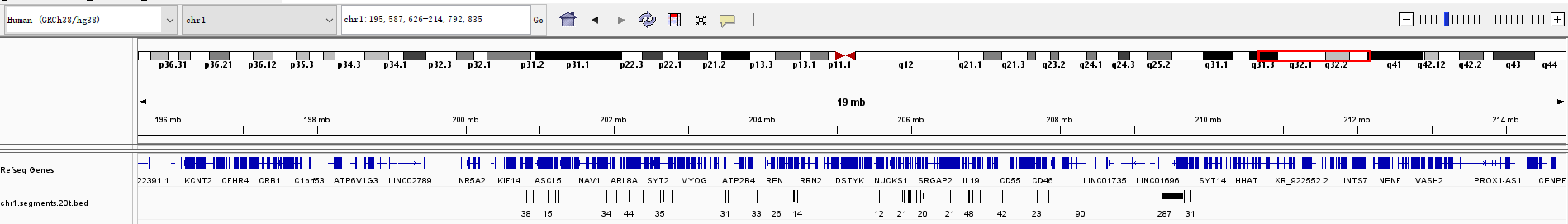
In the file chr1.segments.20.bed, the first column is the read name, you could go to the loci span by that read. Would please also send me the chr1.segments.20.bed file?
This is the chr1.segments.20.bed file chr1.segments.20.txt
The chr1.segments.20.bed file contains around 900 segments, but your log file showed that SVision writes 11170 segments to this file.
I made a test on chr1 of sample HG00514 with the latest code at GitHub, here is the log file I have. The number of segments saved in the output files should be the same as the one showed in the log file.
Please send me your full log file. Thanks!
Here is the lastest full log file. Please take a look at it, Thanks! SVision_221101_212544.log
For src/output_clusters.py, please uncommented the code in line 91 and change it to logging.info('[Debug] Write {0} segments to: {1}'.format(len(all_segs), chr_segments_out_file). You could run SVision in your conda environment via python /path/to/SVision/SVision and please add -c chr1. I need to see the full process in the log file.
Thanks!
Hi,
I'm kind of having a similar issue here. SVision finished segments generation for the whole genome (~56x HiFi, HG002) in around an hour (threads=5), and then stuck there for one day without any new outputs or error logs.
One thing I noticed was that SVision identified much more segments in the windows on hs37d5 compared to other chromosomes which usually have 1~3k segments per window. Does this cause the issue mentioned above?
2023-07-10 16:38:44,514 [INFO ] Processing hs37d5:0-10000000, 15382 segments write to: hs37d5.segments.0.bed 2023-07-10 16:39:20,107 [INFO ] Processing hs37d5:10000000-20000000, 44541 segments write to: hs37d5.segments.1.bed 2023-07-10 17:01:46,751 [INFO ] Processing hs37d5:20000000-30000000, 154348 segments write to: hs37d5.segments.2.bed 2023-07-10 17:08:25,867 [INFO ] Processing hs37d5:30000000-35477943, 22292 segments write to: hs37d5.segments.3.bed
Another question is that, will SVision produce same SV results for chr1 when run with and without -c chr1? I'm considering parallelizing SVision by submiting separate jobs for each chromosome, but kind of afraid that this may give different results
Thank you
hello,
I met this problem when running SVision, too. Do you solve this problem? Addtionally, I want to ask how long time the mission is finished
Thank you for your reply.
@xuexingwangzhedishitian What is the coverage of your data? I also recommend to split your bam file into different chromosome.
FYI. I am working on a new version which is expected to release this month or next.
Thank you for your reply. It is helpful for my job!
My data is ONT data with 40X coverge. Expect the new version
@xuexingwangzhedishitian I would say this method is even slower on ONT data. I highly recommended HiFi data for current version, since the encoded image is really sensitive to ambiguous alignments.
@jiadong324 Thank you for your answer. It's very useful for my work!
Looking forward to the release of the new version of SVision.