Four question about the training model on Lizard dataset with hover_net-conic code
Hello vqdang, I am a student who is learning your code<hover_net-conic> but meeting some question. During the process of learning your code, I meet with 4 questions. Questions are as follows.All the result of pictures are generated by <hover_net-conic> code.
- there are some differents between hover_net-master(MASTER) and hover_net-conic(CONIC), which is CONIC doesn't have inference code. So I transfer run_infer code from MASTER to CONIC, I don't know is it ok to do this.
- As shown in the picture, my trained model doesn't segment lymphocytes very well.

- As shown in the picture, my trained model doesn't classify lymphocytes very well, it recognize lymphocytes as epithelial cells
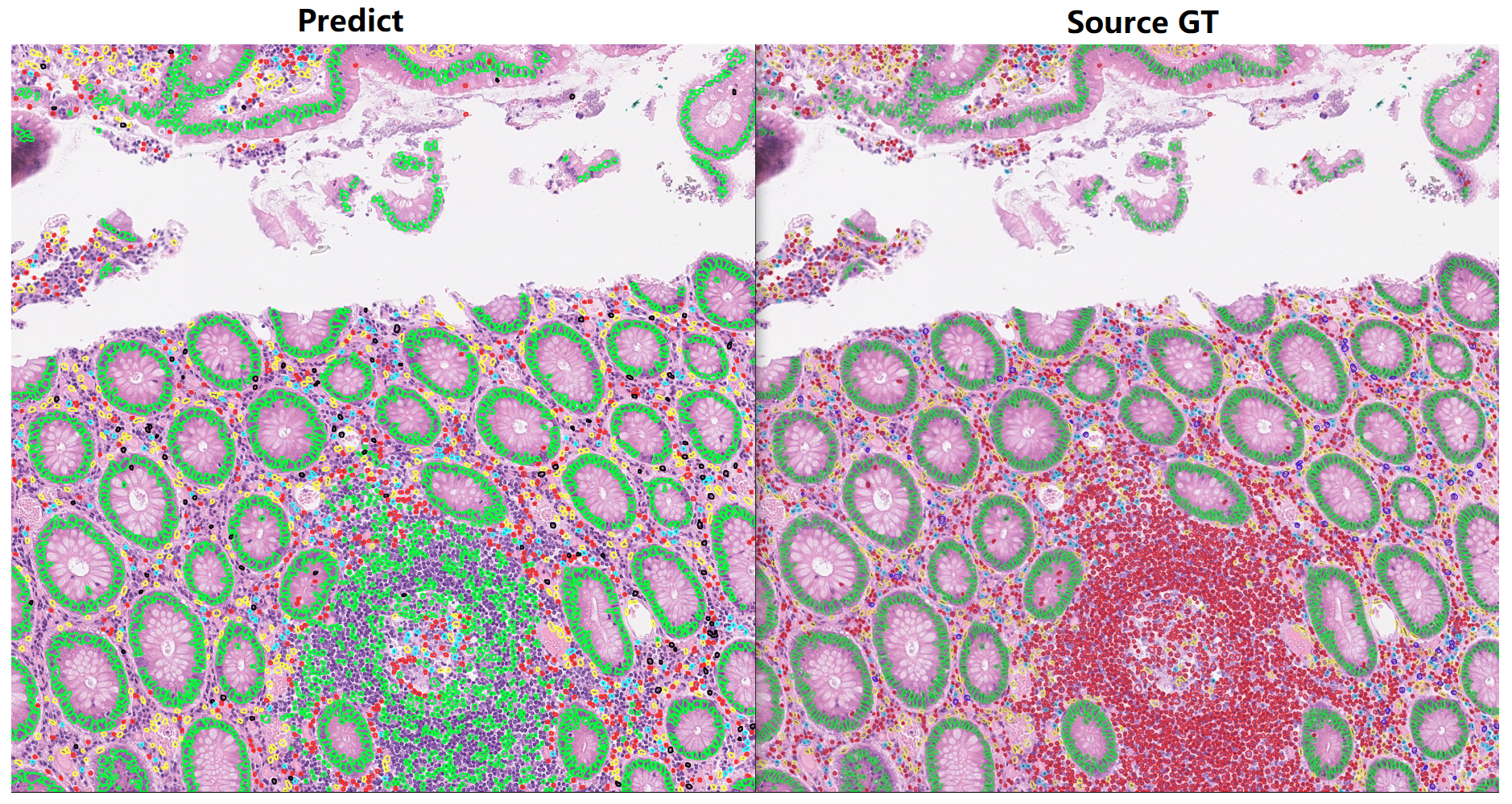
- I find that info from debug.log , infer-valid-tp_dice_1 and infer-valid-tp_dice_5, are consistently 0 on the valididation set. And funcion "remove_small_objects" keeps popping prompts during the running process.

I am looking forward to your reply and guidance! Besides, is it possible for you to share your checkpoint to me to make a inference on the Lizard dataset?
Hi, you can refer to this https://github.com/TissueImageAnalytics/CoNIC/tree/docker-template/conic_baseline to use the inference code. The above page also contains the pretrained HoVerNet on Lizard for the CONIC challenge.
If you are not using the data prepared for CONIC, maybe #4 happen due to heavy class imbalance in your training portion. A larger training batch size also helps.
Hi, you can refer to this https://github.com/TissueImageAnalytics/CoNIC/tree/docker-template/conic_baseline to use the inference code. The above page also contains the pretrained HoVerNet on Lizard for the CONIC challenge.
If you are not using the data prepared for CONIC, maybe #4 happen due to heavy class imbalance in your training portion. A larger training batch size also helps.
Thanks for your reply. Your suggestion is helpful for #4 which was truely caused by the batch size. Is there any other suggestions about questions #2 and #3? Expecially about #3.
My Hyperparameter are as follow, only change the loader_kwargs params. optimizer_kwargs: 'lr' : 1.0e-4 'betas' : [0.9, 0.999] 'weight_decay' : 0.0
loader_kwargs: train: { 'batch_size' : 2, 'nr_procs' : 16} infer: { 'batch_size' : 4, 'nr_procs' : 8}
model_kwargs: num_types: 7 # conic pretrained_backbone: 'pretrain/resnet50-0676ba61.pth'
I think I may know what is wrong then now that I reread your comment.
So I transfer run_infer code from MASTER to CONIC, I don't know is it ok to do this.
CoNIC model and data were designed for 20x dataset. This repository was designed for 40x. Thus, the predictions will be garbage. The conic repository actually supports the inference code. Its in another branch and I guess it is a bit obscure if you didn't follow the Challenge.
https://github.com/TissueImageAnalytics/CoNIC/tree/docker-template/conic_baseline
I think I may know what is wrong then now that I reread your comment.
So I transfer run_infer code from MASTER to CONIC, I don't know is it ok to do this.
CoNIC model and data were designed for 20x dataset. This repository was designed for 40x. Thus, the predictions will be garbage. The conic repository actually supports the inference code. Its in another branch and I guess it is a bit obscure if you didn't follow the Challenge.
https://github.com/TissueImageAnalytics/CoNIC/tree/docker-template/conic_baseline
I got it.
Thus, the predictions will be garbage.
It means that run_infer code from https://github.com/vqdang/hover_net is not suitable for Lizard dataset(20x) directly. Besides, the website below (https://github.com/TissueImageAnalytics/CoNIC/tree/docker-template/conic_baseline) contains suitable code for Lizard dataset inference. is that right?