cant reproduce the simple Nuclear-Application notebook
Describe the bug
when running the notebook on your data (not changing a single line of code), encountering the following error:
ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 96 from C header, got 88 from PyObject
To Reproduce Steps to reproduce the behavior:
- Run from deepcell.datasets.tracked import hela_s3
- Enter '....'
- See error deepcell_toolbox/compute_overlap.pyx in init deepcell_toolbox.compute_overlap()
ValueError: numpy.ndarray size changed, may indicate binary incompatibility. Expected 96 from C header, got 88 from PyObject
Expected behavior the notebook should run like it does in the github example
Screenshots If applicable, add screenshots to help explain your problem.
Desktop (please complete the following information):
- OS: macOS, Windows
- Version 11.6, 11
- Python Version 3.9
Additional context Add any other context about the problem here.
Encountering the same:
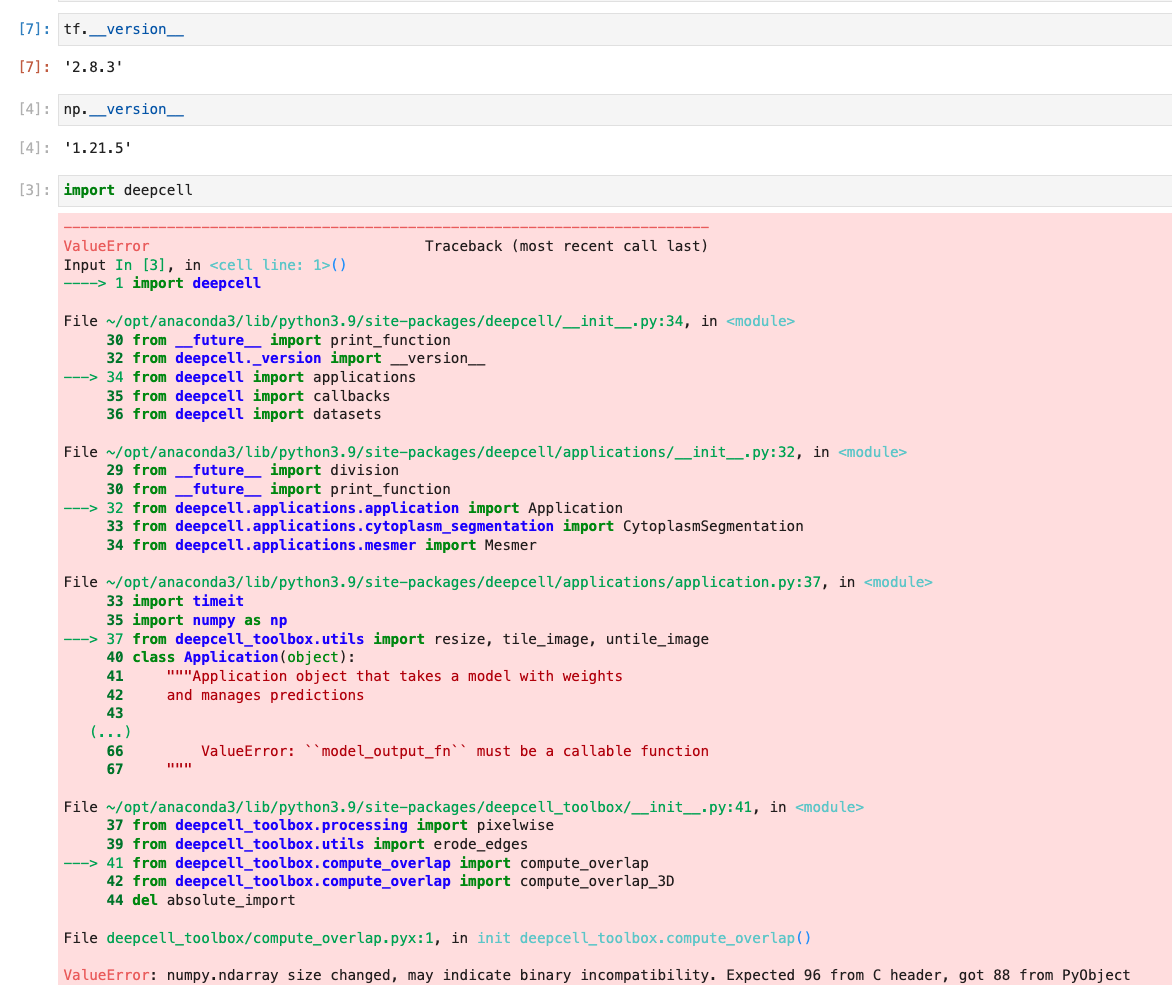
OS: macOS numpy: 1.21.5 tensorflow: 2.8.5 Python: 3.9
Has this issue been resolved? I am facing a similar problem as well.
Thanks for reporting - I suspect this is related to the Cython 3.0 release. We'll try to get a patch out ASAP. In the meantime, if you are running in a locally-managed environment, you can try installing Cython<3 when you install deepcell-toolbox.
I am running it on a cloud based linux machine. I thought deep cell-toolbox is installed when I install the deepcell-tf pipeline using docker. Am I right or would I need to install it separately? If so could you help me with that?
This should be fixed (at least the numpy/cython issue) with the latest 0.12.7 release.