genomepy
 genomepy copied to clipboard
genomepy copied to clipboard
-p localfile
Is there a way of installing genomes from a local file rather than from a URL? In this case the command would be something like
genomepy install -p localfile /path/to/local/directory/genomefile.fa.gz
Or alternatively
genomepy install -p url file:///path/to/local/directory/genomefile.fa.gz
Hey David,
There is! genomepy install -p local /path/to/local/directory/genomefile.fa.gz
Check out genomepy install --help for additional details (e.g. how to also install your own GTF)
Huh, so there is. Is there documentation for this? I don't see that with genomepy install --help
Most install settings are universal, but there is one option specifically for local annotations:
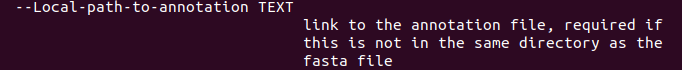
There is currently no additional documentation for the local provider. You're the second person who asked (and on the same day!), so I'll add some when I have the time.
I have a add on question. We are hosting a local mirror of the UCSC genome browser with our own mysql server and web interface. Is there a way of substituting the mysql host for UCSC?
No. Unfortunately, making a generic SQL provider is a bit more difficult as there are at least 2 SQL variants (UCSC is using MariaDB for instance). I also don't see how that would be easy to use (but I could be wrong, since this was my only attempt at SQL so far!).
If you have ideas, I'd happily accept a PR :)