Tali Mazor
Tali Mazor
Probably the same as this earlier issue: https://github.com/cBioPortal/datahub/issues/1515
Good idea @inodb !
Nice addition @BasLee ! I can't test the actual functionality in the public portal, but as @inodb suggested, the separator should not be present if there are no reference genome...
I think adding the M value as a second profile makes a lot of sense
Oh! That makes sense. Using the case lists sounds like a good interim solution until we do the SV switch. Thanks @jjgao @kalletlak
I thought we had put a temporary fix in place, but public genie portal is showing 100% of samples with fusions again: 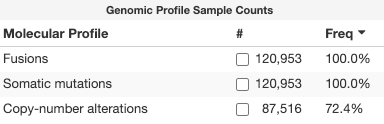
@ritikakundra @jjgao I'm still seeing this issue in the public v11 release. Is there any progress on getting those case lists for GENIE?
This is still an issue in current GENIE v12 release
@ritikakundra hmmm ok - can you re-do for this latest version? and what is the fix? we'll need to do it for genie in our internal portal also.