msmc2
 msmc2 copied to clipboard
msmc2 copied to clipboard
MSMC2 -I Range violation
Dear @stschiff I want to do population analysis using MSMC2, and I generate a combined input file with eight haplotypes like this (generate by 4 sample form 2 population) MayiVsCH_chr10.txt
and my command line was:
${MSMC}/msmc2 -t 15 -p 1*2+25*1+1*2+1*3 -o $OUTDIR/CH.msmc2 -I 0,1,2,3 $INDIR/MayiVsCH_chr*.txt
${MSMC}/msmc2 -t 15 -p 1*2+25*1+1*2+1*3 -o $OUTDIR/Mayi.msmc2 -I 4,5,6,7 $INDIR/MayiVsCH_chr*.txt
when I run this command, it reported an error. the log was, core.exception.RangeError@model/psmc_hmm.d(124): Range violation
??:? _d_arrayboundsp [0x5dcab1] ??:? void model.psmc_hmm.PSMC_hmm.runForward() [0x55b540] ??:? std.typecons.Tuple!(double[][], double[][2], double).Tuple expectation_step.singleChromosomeExpectation(in model.data.SegSite_t[], ulong, in model.propagation_core.PropagationCore) [0x590497] ??:? int expectation_step.getExpectation(in model.data.SegSite_t[][], model.psmc_model.PSMCmodel, ulong, ulong).__foreachbody5(ulong, ref const(model.data.SegSite_t[])) [0x5902af] ??:? void std.parallelism.ParallelForeach!(const(model.data.SegSite_t[])[]).ParallelForeach.opApply(scope int delegate(ulong, ref const(model.data.SegSite_t[]))).doIt() [0x5468ce] ??:? void std.parallelism.run!(void delegate()).run(void delegate()) [0x5ecbeb] ??:? void std.parallelism.Task!(std.parallelism.run, void delegate()).Task.impl(void*) [0x5ec6b7] ??:? void std.parallelism.AbstractTask.job() [0x61e67a] ??:? void std.parallelism.TaskPool.doJob(std.parallelism.AbstractTask*) [0x5eb10b] ??:? void std.parallelism.TaskPool.executeWorkLoop() [0x5eb281] ??:? void std.parallelism.TaskPool.startWorkLoop() [0x5eb227] ??:? void core.thread.context.Callable.opCall() [0x610060] ??:? thread_entryPoint [0x60fb43] ??:? [0x2af2975bae24] ??:? clone [0x2af2981eb34c]
Can you give me some help? Thank you, bx
I will try and reproduce this, will be in touch.
Thank you for your patient answer!
I am so sorry to bother you. Last day, I found when I use the reference genome of Gullus.gullus(Galgal6), it can not call varient in chromosome 29, like this
 When I delete chr29.txt, it works.
then, When I estimate population separation history, some result values(rCCR) are much higher than 1:
When I delete chr29.txt, it works.
then, When I estimate population separation history, some result values(rCCR) are much higher than 1:
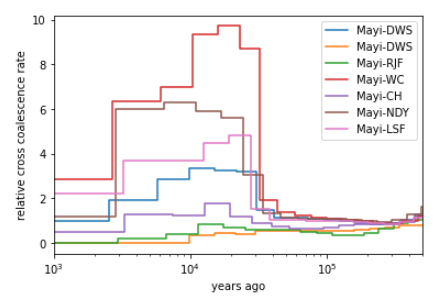
How can I solve this problem? Thank you, bx
OK good. As for interpreting the rCCR: This can happen, perhaps due to bad phasing, or simply because of bad estimates of population sizes. The rCCR is simply the ratio of the cross-coalescence rate and the within-population coalescence rates. So perhaps you should look at all of them and see whether you understand where this comes from. There is no general advice here, I'm afraid
thank you, I will check my dataset and try again
Hello @bxZhang996. I ran into the same problem. After several tries, MSMC2 was still indicating "Range violation" and I noticed I have some multihetsep files with no variants. Did you find a way around it? I'm working only with estimations of population size, if this changes something. Thank you.
I don't understand. If you have empty multihetsep files, you need to obviously exclude them, because they provide no information. Or does that not solve the issue?