R fatal error when calling pairs
Summary:
Calling pairs() on two different models has resulted in 2 R fatal errors/R sessions aborted.
Description:
I created the models with brm() in the BRMS package. I received warnings that there were many (~200) divergent transitions with the first model that crashed and just 1 divergent transition the second time it crashed. I have successfully run the pairs() function once but there were no red dots in it which I expected based on my reading of what the pairs() function does. The other time I managed to get a plot, the plot was broken.
Reproducible Steps:
Here is the data I used: SampleData.xlsx
And the code I used for the second time it crashed:
prior<-get_prior(formula = bf(value ~ 1 + allowed_vote + (1|Participant) + (1|variable))+
lf(disc ~ 0 + allowed_vote, cmc = FALSE),
data = data,
family = acat("probit"))
fit_G3_withPriors<-brm(
formula = bf(value ~ 1 + allowed_vote + (1|Participant) + (1|variable))+
lf(disc ~ 0 + allowed_vote, cmc = FALSE),
data = data,
family = cumulative("probit"),
save_all_pars = TRUE,
prior = prior
)
pairs(fit_G3_withPriors)
Current Output:
The message when crashing with a cumulative model with unequal variances: 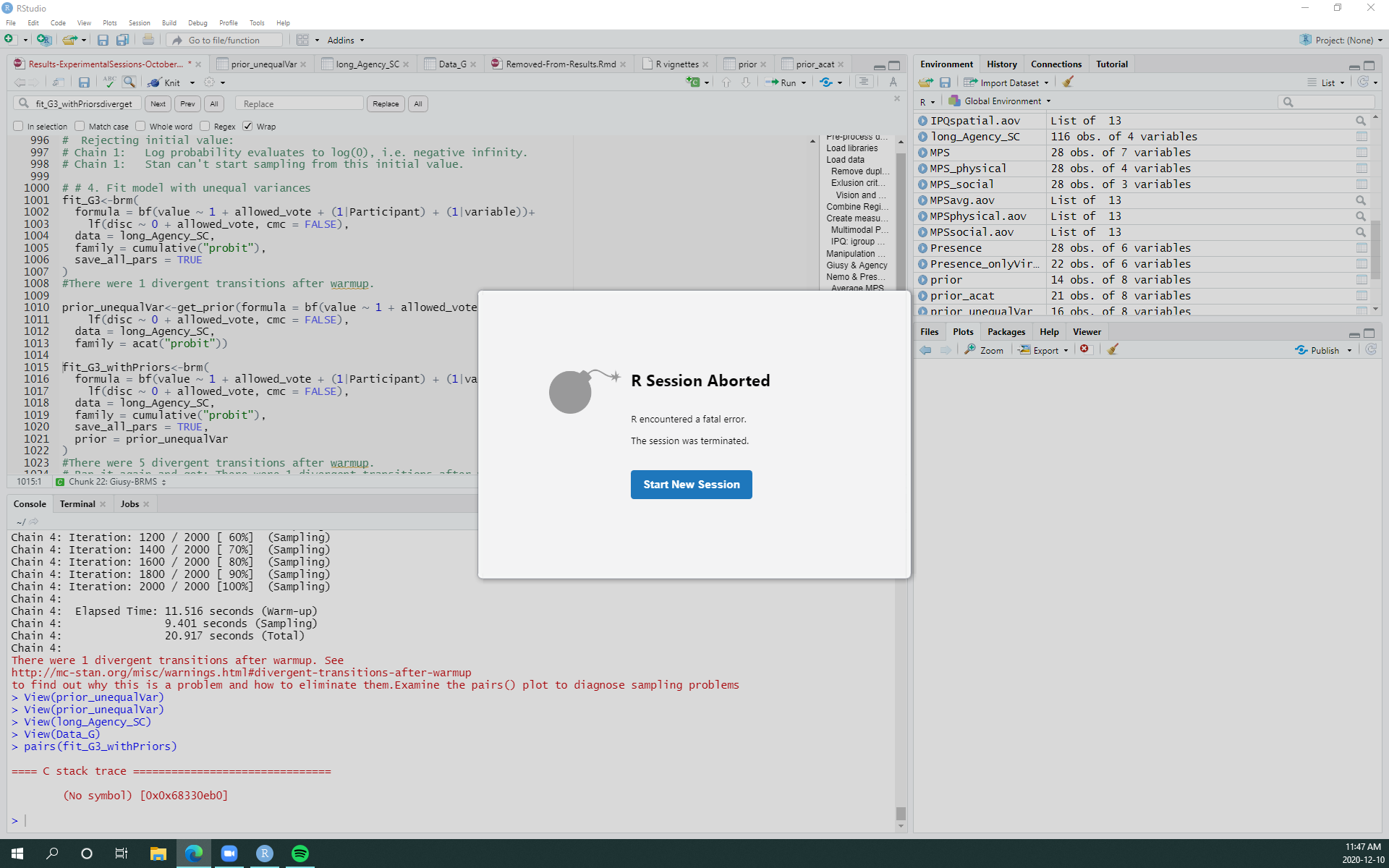
The plot I got that was clearly broken (from running adjacent category model with category-specific effects):

The plot I got when pairs() actually ran with a cumulative model with equal variances:

Expected Output:
The pairs plot is supposed to show divergent transitions with red points but even when pairs ran, I don't see any red points in the pairs() plot (though I believe in that case there was only one divergent transition).
Versions:
rstan_2.21.2 brms_2.13.5 R version 4.0.2
And the call that crashed before, when re-run after restarting R provided this plot:

Hi Dana, sorry about the delay in getting to this. I'm able to run your code above on Windows with no crashes. The main difference I see that I'm using the github version of brms. Can you try installing that and trying your code again? Installed via:
if (!requireNamespace("remotes")) {
install.packages("remotes")
}
remotes::install_github("paul-buerkner/brms")
If you still have crashes, then I'd recommend installing cmdstanR: https://mc-stan.org/cmdstanr/articles/cmdstanr.html, and then running your brms models with the option backend="cmdstanr", since that tends to be more stable.