CRPS and SCRPS
Addresses #136. Adds functions to compute crps and scrps and their loo versions. Happy to hear feedback!
Some math:
CRPS score
The CRPS for forecast CDF $F$ is $$\mathrm{crps}(F,y)=E_X|X-y|- \frac{1}{2}E_{X,X^\prime}|X-X^\prime|.$$ where $X$ and $X'$ are two independent copies from $F$.
Using these predictive draws $z_{m1}, \dots, z_{mS}$, we can compute the crps of the model on the $m$-th datapoint by
$$ \widehat {\mathrm{crps}}{m}= \widehat {\mathrm{crps}}(z_{m1}, \dots, z_{ms}, z_{m})= \frac{1}{S} \sum_{s=1}^S |z_{ms}-z_{m}| - \frac{1}{2S^2} \sum_{s, s'=1}^S |z_{ms}- z_{ms'}|.$$
Fast LOO approximation
For each draw $\theta_s$ from posterior $p(\theta \vert y)$, we simulate a prediction $$\tilde y_{1s}, \tilde y_{2s}, \dots, \tilde y_{ns}$$ from the pointwise sampling distribution $p(y \vert \theta)$.
Fix a data point $m$, we need to compute the LOO-IS ratios and ``product LOO-IS ratio''
$$r_{s} = \frac{ p^{-1}(y_m | \theta_{s} )} {\sum_{j=1}^S p^{-1}(y_{m} | \theta_{ j}) } $$
$$ r_{ss^\prime} = \frac{ p^{-1}(y_{m} | \theta_{s} ) p^{-1}(y_{m} | \theta_{s^{\prime}} )} {\sum_{j, j^\prime=1}^{S} p^{-1}(y_{m} | \theta_{ j}) p^{-1}(y_{m} | \theta_{j^{\prime}} )} $$
We estimate the pointwise crps score by
$$
\widehat {\mathrm{crps}}{m}^{\mathrm{loo}} = \sum{s} r_{s} \vert \tilde y_{ms}-y_{m} \vert -
\frac{1}{2} \sum_{s, s'=1}^S r_{ss^\prime} \vert \tilde y_{ms}- \tilde y_{ms'} \vert
$$
The second term needs to run PSIS on the product ratios.
Jensen inequality
These two schemes are different: $$E_{\theta} E_{ x_1, x_2 \vert \theta } { abs( x_1 - x_2 ) } \neq E_{\theta_1, \theta_2} E_{ x_1 \vert \theta_1 x_2 \vert \theta_2 } { abs( x_1 - x_2 ) }$$
I did RHS in my newer pull request.
Thanks @yao-yl! I compared the crps with the old and suggested version to the one from scoringRules crps_sample-function. With the old implementation, the correlation is close to 1 but there is a small bias (~0.006). When the shuffling is added, estimates are drastically different:
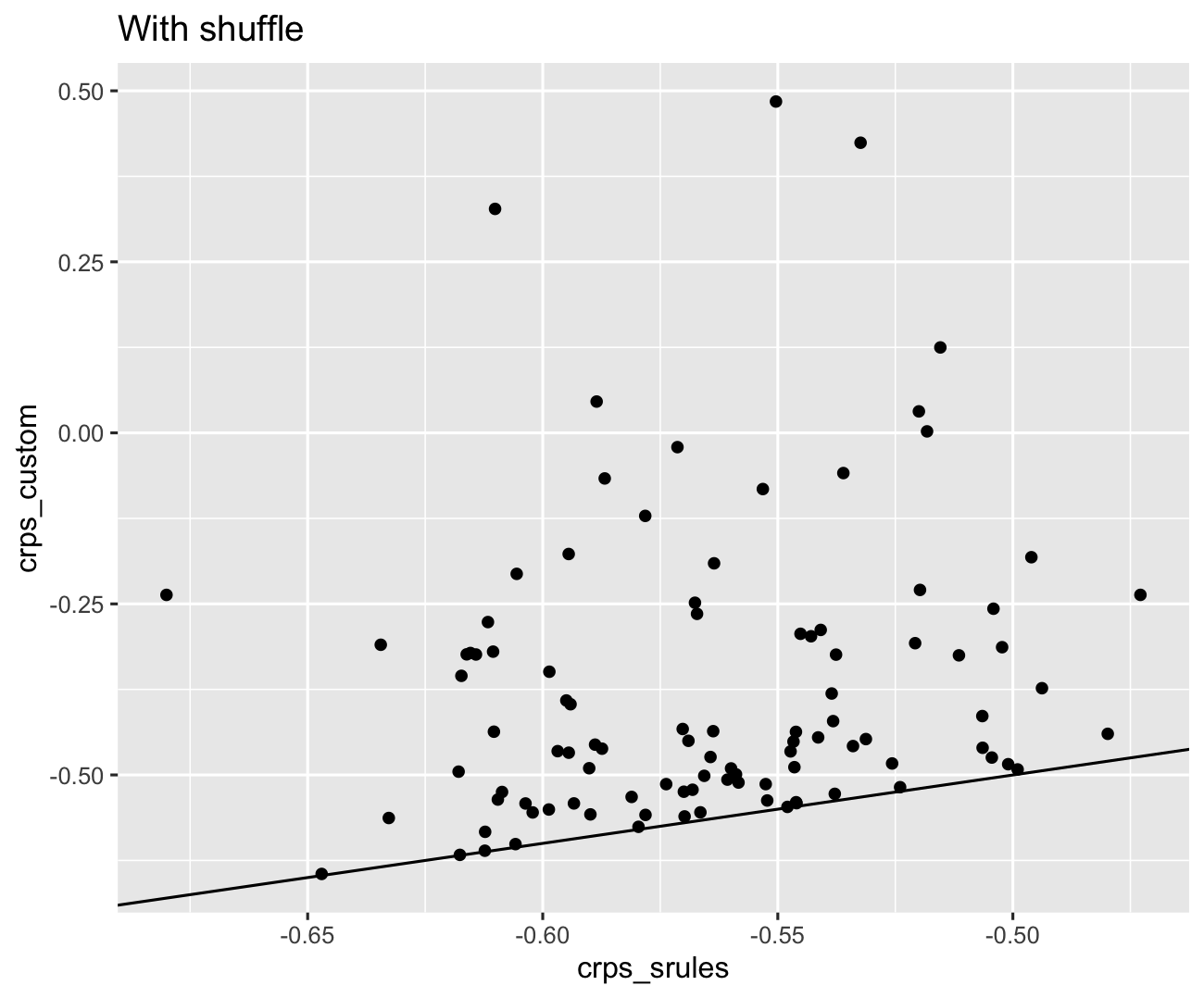 It is not yet clear to me why this happens.
It is not yet clear to me why this happens.
library(rstanarm)
library(ggplot2)
crps_srules <- numeric()
crps_custom <- numeric()
for(i in 1:100) {
x <- rnorm(100)
y <- rnorm(1) + x * rnorm(1) + rnorm(100)
fit <- stan_glm(y ~ x, data = data.frame(y, x), refresh=0)
crps_srules[i] <- mean(-scoringRules::crps_sample(y, t(posterior_predict(fit))))
crps_custom[i] <- crps(posterior_predict(fit), posterior_predict(fit), y)$estimate[1]
}
cor(crps_srules, crps_custom)
qplot(crps_srules, crps_custom) + geom_abline()
@LeeviLindgren I might have misunderstood the shape in your original code:
crps.default <- function(x1, x2, y)
I thought it is point-wise crps, with x1 x2, length S vector and y is a scaler. So I use x2 <- x2[shuffle] to shuffle simulation draws.
But now I realize that you allow y to be a vector. So the correct shuffle should be
x2 <- x2[shuffle, ]
The data (example) index should not be shuffled. We need to take care to of more complex dimensions.
Re bias: the bias will be very small if the bayesian posterior is concentrated (in the big n limit). The bias is bigger is posterior variance of parameters is bigger. Try this example on pointwise CRPS
theta=rnorm(1000,0,2) # parameter inference
y=1 # observed data, only one example
y_pred1=rnorm(theta) # posterior_predict 1
y_pred2=rnorm(theta) # posterior_predict 2
Yeah, I see your point, I should have noticed that! The code should be more explicit about the dimensions. I will modify the methods a bit so that the function works in both of the following cases:
-
x1andx2are vectors of lengthS, andyis a scalar. -
x1andx2are matrices of sizeS x n, andyis a vector ofnobservations.
@LeeviLindgren the separation for the matrix and vector looks good. One more question: when computing Exx, in theory we can average over all permutations to reduce variance (In the current version, we only compare one possible permutations x1, and x2[shuffle]). If we do generate more permutations, the variance will be reduced for Exx, while Exy has a fixed variance term. On the other hand, the estimate of Exx could have a higher variance to begin with because it involves two random copies.
Would it be a good idea to add this permutation times as the function input (at this moment it is 1): computing EXX multiple times and taking the average in crps and loo_crps. It is a further research topic to how to choose the reasonable number of permutations to balance the variance of Exx and Exy.
@LeeviLindgren the separation for the matrix and vector looks good. One more question: when computing Exx, in theory we can average over all permutations to reduce variance (In the current version, we only compare one possible permutations x1, and x2[shuffle]). If we do generate more permutations, the variance will be reduced for Exx, while Exy has a fixed variance term. On the other hand, the estimate of Exx could have a higher variance to begin with because it involves two random copies.
Would it be a good idea to add this permutation times as the function input (at this moment it is 1): computing EXX multiple times and taking the average in crps and loo_crps. It is a further research topic to how to choose the reasonable number of permutations to balance the variance of Exx and Exy.
Did these questions/suggestions from @yao-yl ever get resolved?
Trying to figure out if this is ready or if we're still waiting on some changes.
Sorry for the late reply! @jgabry I implemented the permutation option suggested by @yao-yl. I did some checks today and fixed a minor bug and from my side, it is good for reviewing.
I'm finally getting ready to prepare another loo release in order to fix a test failure for CRAN, but we can also include this PR. @LeeviLindgren the changes look good to me, thanks! I have a few small changes to suggest for the documentation, but I can do those after merging this.
@avehtari Are you good with this? If so, I think we're ready to merge it.
I'm finally getting ready to prepare another loo release in order to fix a test failure for CRAN, but we can also include this PR. @LeeviLindgren the changes look good to me, thanks! I have a few small changes to suggest for the documentation, but I can do those after merging this.
@avehtari Are you good with this? If so, I think we're ready to merge it.
Also @LeeviLindgren could you add me as a collaborator in your fork of the loo repository? I thought it would let me push to this PR but it's not letting me. I guess you might need to add me as a collaborator in your repository settings.
As far as I know, this is ok
@jgabry you should be now a collaborator on my fork.
Great, thanks! I might make a few small changes before merging, but in general this PR looks good. Thanks again @LeeviLindgren. I will also add you to the package DESCRIPTION file as a contributor to the package.
Codecov Report
Merging #203 (0ec8c8e) into master (0fce33b) will decrease coverage by
0.09%. The diff coverage is90.00%.
:exclamation: Current head 0ec8c8e differs from pull request most recent head 1db44b3. Consider uploading reports for the commit 1db44b3 to get more accurate results
@@ Coverage Diff @@
## master #203 +/- ##
==========================================
- Coverage 93.48% 93.40% -0.09%
==========================================
Files 29 30 +1
Lines 2715 2773 +58
==========================================
+ Hits 2538 2590 +52
- Misses 177 183 +6
| Impacted Files | Coverage Δ | |
|---|---|---|
| R/crps.R | 89.65% <89.65%> (ø) |
|
| R/E_loo.R | 98.86% <100.00%> (ø) |
|
| R/psis.R | 93.68% <100.00%> (ø) |
:mega: We’re building smart automated test selection to slash your CI/CD build times. Learn more