bayesplot
 bayesplot copied to clipboard
bayesplot copied to clipboard
feature request: allow changing the labels of the legends
This is an idea that came up in the discussion of #73
The point is to let users change the labels of the legend in bayesplot objects easily, possibly by setting them as arguments in the plot creation. This could for instance be helpful to plot the pp_check for two models and then label y_rep as model Aor model B, respectively.
I mainly used pp_check() until now but I guess this might be usable for other plotting possibilites as well.
@jgabry wrote in #73:
But another option would be to add additional arguments to all of the bayesplot plotting functions so that user's can specify the names to be used in the legend at the same time they create the plot. For example, it could look something like this maybe:
my_legend <- list(y = "emp. data", yrep = "model predict.") ppc_bars(y, yrep, ..., legend_labels = my_legend)That way the legends could be created with those labels instead of having to change them after the fact. I'd need to figure out the best way to implement that (it would vary depending on the plotting function) but that should be much easier to do.
Thanks for opening the issue. This is officially on the to-do list now!
On Wed, Mar 1, 2017 at 6:19 AM silberzwiebel [email protected] wrote:
This is an idea that came up in the discussion of #73 https://github.com/stan-dev/bayesplot/issues/73
The point is to let users change the labels of the legend in bayesplot objects easily, possibly by setting them as arguments in the plot creation. This could for instance be helpful to plot the pp_check for two models and then label y_rep as model Aor model B, respectively. I mainly used pp_check() until now but I guess this might be usable for other plotting possibilites as well.
@jgabry https://github.com/jgabry wrote in #73 https://github.com/stan-dev/bayesplot/issues/73:
But another option would be to add additional arguments to all of the bayesplot plotting functions so that user's can specify the names to be used in the legend at the same time they create the plot. For example, it could look something like this maybe:
my_legend <- list(y = "emp. data", yrep = "model predict.") ppc_bars(y, yrep, ..., legend_labels = my_legend)
That way the legends could be created with those labels instead of having to change them after the fact. I'd need to figure out the best way to implement that (it would vary depending on the plotting function) but that should be much easier to do.
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/stan-dev/bayesplot/issues/75, or mute the thread https://github.com/notifications/unsubscribe-auth/AHb4Q1AGq624obkz8xvzoh0kQxBAUJ_eks5rhVRFgaJpZM4MPhxq .
Just stumbling over this again. The work-around for changing the text of the legend still works but now I wanted to change the text of facet strings. In my concrete example I want to contrast the outcome for a boolean factor and I'm just seeing "TRUE" and "FALSE" as facet labels. However, manual context-sensitive strings would be way more intuitive for the plot. Should I update the the title of this issue to include other labels than the legend, too, or rather open a new feature request?
@silberzwiebel Thanks for following up on this!
I would much rather add helper functions for this to modify these things after the plot is created (e.g., like these other ones we have for stuff like changing axis titles) than add many more arguments to the plotting functions, but as far as I know it's not possible to change the legend or facet labels after the fact. @tjmahr Any clever ideas in this regard? Do you know of a way to edit the created ggplot object to change this stuff (maybe messing with ggplot_build)?
In other words, if we can allow this by adding a few helper functions it would be so much better than having to add code for this to each of the plotting functions individually.
Helper functions sound great. What you wrote the other year for changing the legend strings was:
p$scales$scales[[1]]$labels <- "emp. data" p$scales$scales[[2]]$labels <- "model predict."
This happens after the plot is already built (and works fine), so it could go into a helper function. But I'm entirely unsure and unexperienced whether these scales are "stable" in the sense that they will always correspond to the legend and whether it is possible to change other strings like the facet labels in this way.
This happens after the plot is already built (and works fine), so it could go into a helper function. But I'm entirely unsure and unexperienced whether these scales are "stable" in the sense that they will always correspond to the legend and whether it is possible to change other strings like the facet labels in this way.
Yeah, what's tricky about that is the order of scales may be different for the different plots. So it would be great to have a function to use like
p + legend_labels(c("empirical data", "model prediction"))
but my hunch is that legend_labels would have to have lots of different logic inside to deal with the different possible plots. Maybe possible though.
Regarding the facet labels, can you give more details about what you want to do? It should actually already be possible to change the facet labels for many of the plotting functions, but you have to use a ggplot labellerfunction as described in ?ggplot2::labeller. For example, here's how you would do it for ppc_bars_grouped:
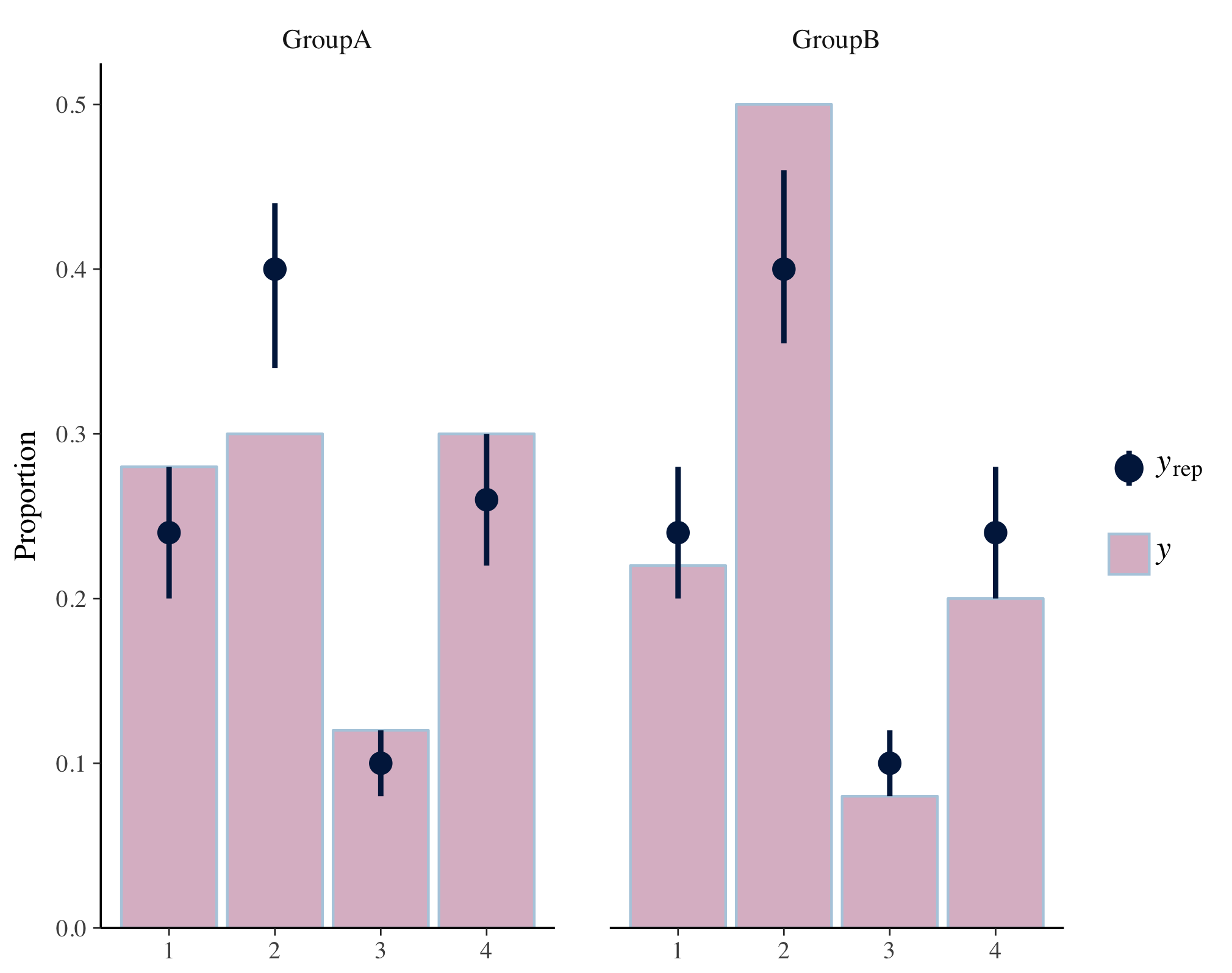
# simulate some data
f <- function(N) {
sample(1:4, size = N, replace = TRUE, prob = c(0.25, 0.4, 0.1, 0.25))
}
y <- f(100)
yrep <- t(replicate(500, f(100)))
dim(yrep)
group <- gl(2, 50, length = 100, labels = c("GroupA", "GroupB"))
# facet labels default to labels of 'group' (in this case GroupA, GroupB)
p <- ppc_bars_grouped(y, yrep, group, prob = 0.5, freq = FALSE)
plot(p)
Now we can change the facet labels to "Banana" and "Turtle" instead of "GroupA" and "GroupB":
my_labels <- as_labeller(c("GroupA" = "Banana", "GroupB" = "Turtle"))
p + facet_wrap("group", labeller = my_labels)

Wow, thanks, changing facet labels like this works fine! I had some trouble because my grouping variable has a different name than group but still the first argument of facet_wrap("group", labeller = my_labels) needs to be "group", otherwise ggplot2 will complain. Apart from that: Thanks for the ggplot2 support :)
Is there already a hint to this approach somewhere in bayesplot's documentation and I was too ignorant to find it?
Not too ignorant! It’s not currently in any examples anywhere, but it would
be a welcome addition. For this we could also add a facet_relabel helper
function.
On Mon, Apr 16, 2018 at 4:21 AM kartoffelsalat [email protected] wrote:
Wow, thanks, changing facet labels like this works fine! I had some trouble because my grouping variable has a different name than group but still the first argument of facet_wrap("group", labeller = my_labels) needs to be "group", otherwise ggplot2 will complain. Apart from that: Thanks for the ggplot2 support :) Is there already a hint to this approach somewhere in bayesplot's documentation and I was too ignorant to find it?
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/stan-dev/bayesplot/issues/75#issuecomment-381518741, or mute the thread https://github.com/notifications/unsubscribe-auth/AHb4Q2t97_YWsgg5RMamrlIrQCMu4vyHks5tpFSKgaJpZM4MPhxq .
If you point me to an appropriate location in an appropriate file, I could write some examples lines on how to change the facet labels (although you might be quicker in doing this yourself -- doesn't matter to me).
Right now there’s not one place to put it that makes sense since it applies to any of the ones with facets. It probably makes sense to add a facet_relabel() helper function and then put an example in the documentation in
https://github.com/stan-dev/bayesplot/blob/master/R/bayesplot-helpers.R
How about I do the function and then you can add an example you like to the doc? I could easily add the example, but I like encouraging users to contribute! Maybe you’ll be more likely to suggest another useful addition in the future ;)
Sounds great!
@silberzwiebel I just pushed a branch called "facet_relabel" that has new functions facet_relabel_gg() (do the facet relabeling) and facet_vars() (get info about variable and scales bayesplot used for faceting). There's documentation explaining them too that you can look at for the details.
I added one example using mcmc_trace, but it would great if you can add one for PPC grouped plots too. I made a spot for your example here:
https://github.com/stan-dev/bayesplot/blob/6b86e130d30be773f73d7527a1def46b84800c41/R/bayesplot-helpers.R#L236
To add the example you can fork the bayesplot repo, switch to the facet_relabel branch, add the example, and then submit a PR to the facet_relabel branch (not master). Then I'll merge your PR and make a PR against master to get it into the next release. Let me know if you have any questions about that process and thanks for your willingness to help!
Helper functions sound great. What you wrote the other year for changing the legend strings was:
p$scales$scales[[1]]$labels <- "emp. data" p$scales$scales[[2]]$labels <- "model predict."
This happens after the plot is already built (and works fine), so it could go into a helper function.
For this case, I would suggest using a scale_ function from ggplot2. The example below I update the labels for the color scale:
ggplot(iris) +
aes(x = Sepal.Length, y = Sepal.Width, color = Species) +
geom_point() +
scale_color_discrete(
labels = c(
# old_name = "new name"
setosa = "SETOSA",
versicolor = "VERSICOLOR",
virginica = "VIRGINICA"))
Hah, thanks @tjmahr! Seems obvious in hindsight (especially since I used that method inside of bayesplot) but totally slipped my mind.
On Thu, Apr 19, 2018 at 9:21 AM TJ Mahr [email protected] wrote:
Helper functions sound great. What you wrote the other year for changing the legend strings was:
p$scales$scales[[1]]$labels <- "emp. data" p$scales$scales[[2]]$labels <- "model predict."
This happens after the plot is already built (and works fine), so it could go into a helper function.
For this case, I would suggest using a scale_ function from ggplot2. The example below I update the labels from the color scale:
ggplot(iris) + aes(x = Sepal.Length, y = Sepal.Width, color = Species) + geom_point() + scale_color_discrete( labels = c( # old_name = "new name" setosa = "SETOSA", versicolor = "VERSICOLOR", virginica = "VIRGINICA"))
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub https://github.com/stan-dev/bayesplot/issues/75#issuecomment-382732507, or mute the thread https://github.com/notifications/unsubscribe-auth/AHb4Q1xXfv-A9PFczihemhQeIzGpLigwks5tqI9GgaJpZM4MPhxq .
@jgabry just to let you know that this is still on my list, I' just busy doing other stuff right now (as always ;)).
Thanks, no rush!
@silberzwiebel I was going through open issues and just noticed I hadn't merged your PR (I could have sworn I did that months ago!). So I just finally merged your PR into my facet_relabel branch (not master branch yet). Sorry that took so long. I will now make a PR to master from facet_relabel so @tjmahr can take a look and see if we should get this into a release or not.
Thanks! Actually, I was the one who delayed this by months. I opened the PR just at the end of August, so you did all fine.