ColabFold
 ColabFold copied to clipboard
ColabFold copied to clipboard
colabfold_batch still runs a MSA query even if `--msa-mode single_sequence`
Expected Behavior
No query should be sent to the server if --msa-mode single_sequence
colabfold_batch --num-recycle 6 --msa-mode single_sequence --model-type AlphaFold2-multimer-v2 --recompile-padding 1 ./{fn} .
Current Behavior
Queries are throttled!
2022-04-14 16:47:58,472 Running colabfold 1.2.0 (6b6898514ec50dfc1a46c2c6cfa787319e7289ff)
2022-04-14 16:47:58,485 Found 2 citations for tools or databases
2022-04-14 16:48:06,026 Query 1/1: nLuc-B_BC-cLuc (length 114)
2022-04-14 16:48:08,204 Sleeping for 6s. Reason: RATELIMIT
2022-04-14 16:48:15,699 Sleeping for 7s. Reason: RATELIMIT
2022-04-14 16:48:25,661 Sleeping for 7s. Reason: RATELIMIT
2022-04-14 16:48:34,795 Sleeping for 6s. Reason: RATELIMIT
2022-04-14 16:48:42,303 Sleeping for 6s. Reason: RATELIMIT
2022-04-14 16:48:50,748 Sleeping for 10s. Reason: RATELIMIT
2022-04-14 16:49:09,517 Sleeping for 5s. Reason: RATELIMIT
2022-04-14 16:49:16,348 Sleeping for 9s. Reason: RATELIMIT
2022-04-14 16:49:26,972 Sleeping for 7s. Reason: RATELIMIT
2022-04-14 16:49:35,504 Sleeping for 10s. Reason: RATELIMIT
2022-04-14 16:49:47,474 Sleeping for 10s. Reason: RATELIMIT
2022-04-14 16:49:59,980 Sleeping for 9s. Reason: RATELIMIT
2022-04-14 16:50:10,974 Sleeping for 6s. Reason: RATELIMIT
2022-04-14 16:50:19,813 Sleeping for 7s. Reason: PENDING
Steps to Reproduce (for bugs)
colabfold_batch --num-recycle 6 --msa-mode single_sequence --model-type AlphaFold2-multimer-v2 --recompile-padding 1 ./{fn} .
Your Environment
{
"num_queries": 1,
"use_templates": false,
"use_amber": false,
"msa_mode": "single_sequence",
"model_type": "AlphaFold2-multimer-v2",
"num_models": 5,
"num_recycles": 6,
"model_order": [
3,
4,
5,
1,
2
],
"keep_existing_results": true,
"rank_by": "multimer",
"pair_mode": "unpaired+paired",
"host_url": "https://api.colabfold.com",
"stop_at_score": 100,
"recompile_padding": 1.0,
"recompile_all_models": false,
"commit": "6b6898514ec50dfc1a46c2c6cfa787319e7289ff",
"version": "1.2.0"
}
@martin-steinegger I think there should be a case to handle the single sequence mode of paired and unpaired here: https://github.com/sokrypton/ColabFold/blob/f3f924e4d0acc69ebab7083ca895339976e57f12/colabfold/batch.py#L669
And running with
--msa-mode single_sequence --pair-mode unpaired returns
2022-04-14 17:56:17,482 Query 1/1: nLuc-ABC_B-cLuc (length 154)
2022-04-14 17:56:17,486 Could not generate input features nLuc-ABC_B-cLuc: 'NoneType' object is not subscriptable
Traceback (most recent call last):
File "/home/aljubetic/AF2/CF2/colabfold-conda/lib/python3.7/site-packages/colabfold/batch.py", line 1141, in run
model_type,
File "/home/aljubetic/AF2/CF2/colabfold-conda/lib/python3.7/site-packages/colabfold/batch.py", line 839, in generate_input_feature
paired_msa[sequence_index]
TypeError: 'NoneType' object is not subscriptable
2022-04-14 17:56:17,488 Done
I'll guess I can get arounds this by making a paired+unpaired a3m my self? (I'm just not sure how a custom a3m alignment is passed down) (@sokrypton)
I pushed some change that should fix it. Could you give it a try again please?
Thanks! this fixes the call to the server, but sets paired_a3m_lines = None
Is that the expected behavior. For a single sequence I would expect:
>unpaired
AAAAAAAAAAAAAAAAAAAAAAAAAA-----------------------------------------------
--------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
>paired
AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
>paired+unpaired
AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
AAAAAAAAAAAAAAAAAAAAAAAAAA-----------------------------------------------
--------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
@martin-steinegger Sorry I was just running some old code (so you got a lot of traffic from 193.2.14.82). Should be fixed now.
( PS Is [paired_a3m_lines = None] the correct behavior)
For heterodimers it looks like the coverage is as I would expect:
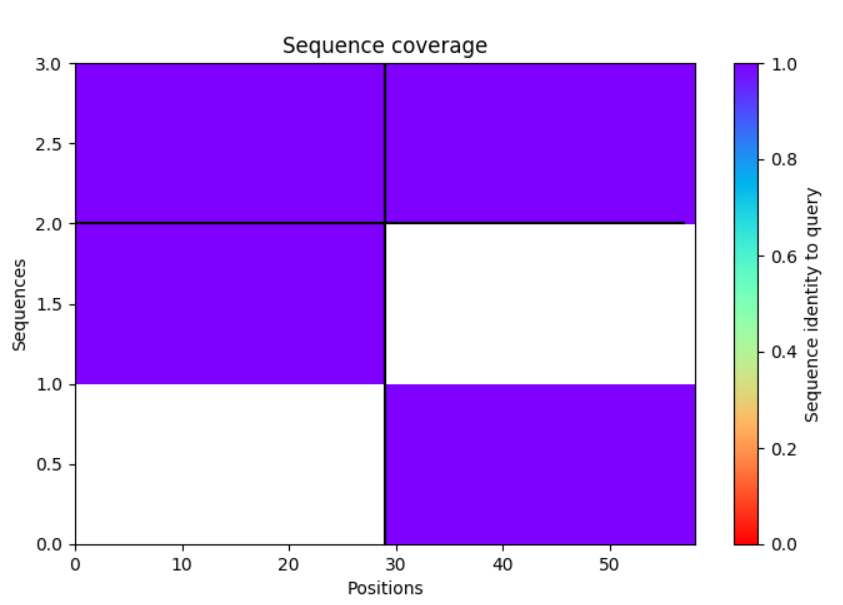 But for homodimers it looks like:
But for homodimers it looks like:
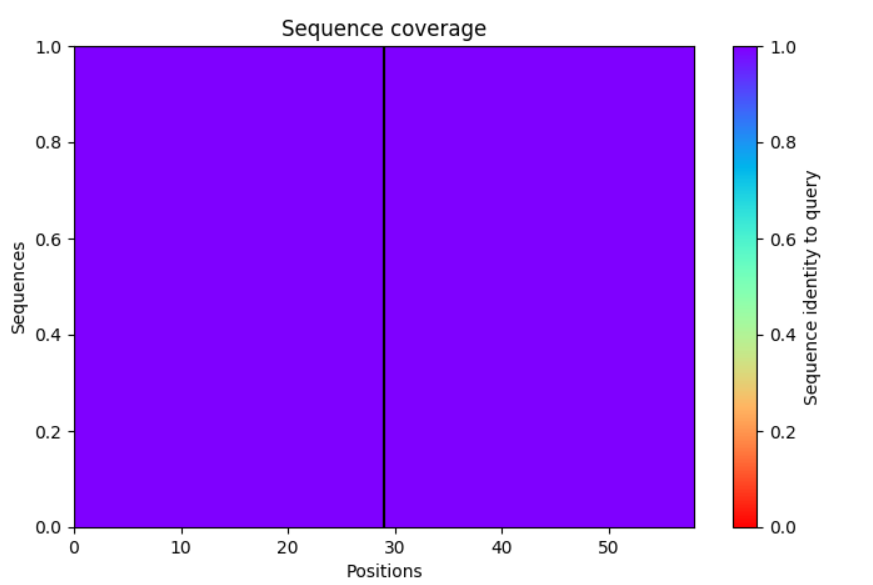
So there is no paired+unpaired.
@sokrypton Is there any advantage to have multiple copies of the same sequence? I think superfold does something similar.
Exmaple:
unpaired AAAAAAAAAAAAAAAAAAAAAAAAAA----------------------------------------------- AAAAAAAAAAAAAAAAAAAAAAAAAA----------------------------------------------- AAAAAAAAAAAAAAAAAAAAAAAAAA----------------------------------------------- AAAAAAAAAAAAAAAAAAAAAAAAAA----------------------------------------------- AAAAAAAAAAAAAAAAAAAAAAAAAA----------------------------------------------- --------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB --------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB --------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB --------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB --------------------------BBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
paired AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB AAAAAAAAAAAAAAAAAAAAAAAAAABBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBBB
For alphafold-multimer putting them on a line, shouldn't be a problem, but for alphafold-ptm... this can be an issue. I'll see if I can fix this in the beta version!
Thanks for the alert.