How to convert logodds explanations to probabilities?
Hey there,
I reviewed this question and I was wondering if I can convert those logodd explanations to probabilities? Essentially, I'd like to see if I can provide insight into how much the probability is influenced by each feature.
To do this, I used the following formula:
1 + 1 / exp(-logodd)
and I would expect that after I convert all of the explanations for a specific prediction to probabilities, the sum should be zero. This ended up not being the case so now i'm a bit stuck.
Any help would be much appreciated.
The #350 thread is a bit old. You can now get SHAP values in probability space directly if you want by using the model_output="probability" option with TreeExplainer. As you discovered, it is not so simple as just running a simple post-transform on the log-odds explanation values.
Thank you for your response, this was very helpful.
I had one follow up question that is related. To double check my shap values for an individual prediction, I took the sum of the shap values and compared it to the output value in my force plot. I'm noticing that these values are slightly off.
Do you know why that might happen? Also, let me know if you'd like me to open up another issue.
Did you add the explainer.expected_value as well? It should always match the difference between the expected model output and the actual output for a given sample.
Here is the code i've used.
explainerModel_prob = shap.TreeExplainer(xgb_model,data = shap.sample(df, 100),model_output='probability')
shap_values_model_prob = explainerModel_prob.shap_values(df)
shap.initjs()
shap.force_plot(explainerModel_prob.expected_value, shap_values_model_prob[5],df.iloc[[0]])
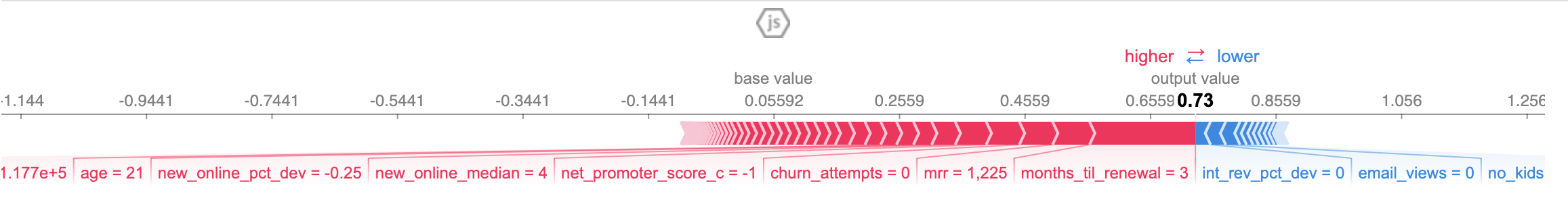
print(np.sum(shap_values_model_prob[5]))
0.67
The output_value in the force plot matches the output probability of my model, its the sum of my shape values that doesn't.
Ah. That's because you need to use explainerModel_prob.expected_value + shap_values_model_prob[5].sum() to match the model output. The SHAP values sum up to the difference between the expected model output and the current model output.
Thanks! So, can you elaborate on how this expected_value is calculated? I looked through your code and saw this piece:
elif data is not None:
try:
self.expected_value = self.model.predict(self.data, output=model_output).mean(0)
When I try to calculate this manually, I get a different value. See below:
explainerModel_prob.expected_value
0.055917
np.mean(xgb_model.predict(shap.sample(df)))
0.02
Could you share a complete example with number? It is hard to know what would have caused the difference in isolation. Perhaps xgb_model.predict is outputting binary labels and not probabilities?
Ah, silly mistake. Thanks for the response. I did have one more question:
I started analyzing the top drivers using SHAP and compared the results using probabilities and log odds. I noticed that the top drivers are different for the same prediction.
Why do you think that would occur? Is that because I'm sampling only 100 data points in the TreeExplainer function? If this difference is expected, how do you suggest I decide which to use for analysis?
A difference in the ordering happens when some features are moving you a long way in long odds space (compared to others), but only moving the model output a short distance in probability space (because you are close to the tails of the logistic). Which one you prefer depends on which units you think are more important. It is very important to go from 0.98 to 0.999 in probability? Then use log odds. If you think it is not important then don't. Depends on the application which makes more sense.
Got it, thanks again. One last question, I also noticed that for a particular prediction, the directionality of a specific driver changes. This seems a bit odd.
Any ideas why that might be?
@slundberg Hey, First, I want to thank you not only for the great tool but also for your responsiveness and explanations as I read and followed quite a few issues here. I am curious, since I played around with the conversions from log-odds space to probabilities about the new model_output='probabilities' feature. As far as I understand, so far the conversion that was used in the tool to visualize the force plot in probabilities space (link='logit') was an approximation that basically transformed the output value and the base value from log-odds to probabilities and then "stretched" the shapely values between them, maintaining the log-odds scale. Now, I see that you introduced the model_output='probabilities' option. Is this also an approximation? And more interestingly, why does it assume feature independence?
Thank you for your help!
@madsthaks That can happen because the average impact of that driver over the background data set can change when we are averaging in log-odds space vs probability (think about large log-odds changes that only change probabilities from 0.99 to 0.999).
@dinaber The link='logit' option to force_plot just makes a non-linear plotting axis, so while the pixels (and hence bar widths) remain in the log-odds space, the tick marks are in probability space (and hence are unevenly spaced). The model_output='probability' option actually rescales the SHAP values to be in the probability space directly. This is a bit of an approximation since it uses the Deep SHAP rescaling approach on the link function after using the exact Tree SHAP algorithm on the margin output of the trees. The reason it only works with "interventional" feature perturbation is that we have to rescale individually per background reference sample, and the "tree_path_dependent" perturbation method can't as easily decompose into a computation on individual background samples. I should note though that 'interventional' does not need to assume feature independence, see https://arxiv.org/abs/1910.13413 for a good overview of why (the interventional option is what they argue for in that paper).
@slundberg Thank you very much for the extensive explanation! I looked at the suggested article. I understand that they suggest using the "interventional" method for marginalization over unused features is the correct way to go to begin with, regardless of whether I want to output in probabilities?
Here is the code i've used.
explainerModel_prob = shap.TreeExplainer(xgb_model,data = shap.sample(df, 100),model_output='probability') shap_values_model_prob = explainerModel_prob.shap_values(df) shap.initjs() shap.force_plot(explainerModel_prob.expected_value, shap_values_model_prob[5],df.iloc[[0]])
print(np.sum(shap_values_model_prob[5])) 0.67The output_value in the force plot matches the output probability of my model, its the sum of my shape values that doesn't. Hi, may I know how do you get this work? I receive error like objective is not define. But i already define is at multi:softprob during the model params. Can i see your full code? Hi, nvm. Ijust found out model_outpput='probability' is only for binary classification. Not multiclass. That's y it doesnt work.
@slundberg is there anywhere we can see how model_probability = 'probability' makes the transformation?
@dinaber that's right!
@zakraicik the logistic transform is used in the C++ code. You can trace it's use to see how it is used to rescale the SHAP values individually for each background reference sample: https://github.com/slundberg/shap/blob/aee2310c21383f9462210c616b0a89c9ed4611a2/shap/tree_shap.h#L135-L137
@slundberg thanks. can you just tell me what the margin represents here?
@slundberg Also wondering how to transform shap_values back to probabilities for multiclass DeepExplainer.
For others out there, I realized it says in the documentation:
"By integrating over many background samples DeepExplainer estimates approximate SHAP values such that they sum up to the difference between the expected model output on the passed background samples and the current model output (f(x) - E[f(x)])."
as such (approximately equal)
model.predict(X_val[i]) == np.sum(e.shap_values(X_val[i])) + e.explainer.expected_value.numpy()
Yeah but If your model output is log odds it is not easy to transform log odd contributions for an individual feature into probability contributions for that feature.
Sent from my iPhone
On Mar 10, 2020, at 11:04 AM, Amund Vedal [email protected] wrote:
For others out there, I realized it says in the documentation:
"By integrating over many background samples DeepExplainer estimates approximate SHAP values such that they sum up to the difference between the expected model output on the passed background samples and the current model output (f(x) - E[f(x)])."
as such model.predict(X_val[i]) ≈ np.sum(e.shap_values(X_val[i])) + e.explainer.expected_value.numpy()
— You are receiving this because you were mentioned. Reply to this email directly, view it on GitHub, or unsubscribe.
@slundberg any thoughts on which is better, rescale or reveal cancel rule, when moving to probability space from margin for xgboost models? The Deep Lift paper gives a heuristic explanation for why the reveal cancel rule gives more meaningful results in their view for certain nonlinear operations, but it's pretty hard to find a more detailed justification than the toy problem they argue it's better on.
Hello,
I try to calculate SHAP values in probabilities with shap.LinearExplainer, but it doesn't seem to work.
The code:
explainer = shap.LinearExplainer(model, data, model_output = "probability")
This just provides log-odds outputs.
@ori-katz100 I can't find it in the documentation and it looks like I'm getting log-odds as well. Has support for model_output='probability' dropped?
I am facing the similar issue, model='probability' gives log odds? Is there a implementation to convert the log odds from SHAP to probability contribution for every feature
@dinaber that's right!
@zakraicik the logistic transform is used in the C++ code. You can trace it's use to see how it is used to rescale the SHAP values individually for each background reference sample:
https://github.com/slundberg/shap/blob/aee2310c21383f9462210c616b0a89c9ed4611a2/shap/tree_shap.h#L135-L137
@slundberg Is it possible to modify this transformation according to needs or isn't it that simple? I need explanations in probability space, but slightly modified - probability scores from my model have skewed distribution and I'd like to make it more uniform by applying the logarithmic transformation: log(score + epsilon). I know that it can't be done by just setting model_output="probability" option and then applying the transformation on received explanations. So is there a way to specify your own transformation e.g. here in tree_shap.h to obtain explanations rescaled properly?
When I am trying to get probabilities, I am getting weird error not sure why. ` params = { "eta": 0.2, "objective": "binary:logistic", "subsample": 0.5, "base_score": 0.5, "eval_metric": "logloss", "scale_pos_weight": scale_pos_weight, "seed": 7 }
model = xgboost.train(params, d_train, 200, evals = [(d_val, "eval")], verbose_eval=10, early_stopping_rounds=20)
explainer = shap.TreeExplainer(model, data = shap.sample(X_train, 100), model_output = "probability", feature_perturbation = "interventional") `
Error Trace:
Exception ignored in: 'array_dealloc'
SystemError: <class 'DeprecationWarning'> returned a result with an error set
SystemError Traceback (most recent call last)
/opt/conda/lib/python3.7/site-packages/shap/explainers/_tree.py in init(self, model, data, model_output, feature_perturbation, feature_names, **deprecated_options) 145 self.feature_perturbation = feature_perturbation 146 self.expected_value = None --> 147 self.model = TreeEnsemble(model, self.data, self.data_missing, model_output) 148 self.model_output = model_output 149 #self.model_output = self.model.model_output # this allows the TreeEnsemble to translate model outputs types by how it loads the model
/opt/conda/lib/python3.7/site-packages/shap/explainers/_tree.py in init(self, model, data, data_missing, model_output) 813 self.model_type = "xgboost" 814 xgb_loader = XGBTreeModelLoader(self.original_model) --> 815 self.trees = xgb_loader.get_trees(data=data, data_missing=data_missing) 816 self.base_offset = xgb_loader.base_score 817 self.objective = objective_name_map.get(xgb_loader.name_obj, None)
/opt/conda/lib/python3.7/site-packages/shap/explainers/_tree.py in get_trees(self, data, data_missing) 1528 "value": self.values[i,:l], 1529 "node_sample_weight": self.sum_hess[i] -> 1530 }, data=data, data_missing=data_missing)) 1531 return trees 1532
/opt/conda/lib/python3.7/site-packages/shap/explainers/_tree.py in init(self, tree, normalize, scaling, data, data_missing) 1353 _cext.dense_tree_update_weights( 1354 self.children_left, self.children_right, self.children_default, self.features, -> 1355 self.thresholds, self.values, 1, self.node_sample_weight, data, data_missing 1356 ) 1357
SystemError:
from math import exp #log odds to probs def logit2prob(logit): odds = exp(logit) prob = odds / (1 + odds) return(prob) #shap_values converter def shap_log2pred_converter(shap_values): return([(logit2prob(x)-0.5) for x in shap_values]) #shap_log2pred_converter(shap_values_test[0][1]) if 2 classes 0 class, 1 example This is how you can translate for DeepExplainer shap values, and there is some problem, it seams like force plot is calculating predicted value from shap values so you need to logit back this probabs.
Any conclusion on the model_output='probability' for linearExplainers? I am getting shap values in the log-odds space despite using the argument model_output, do you know if there is support for that? It doesn't look like, according to the documentation...
How to convert interaction shap values into probability?
If there is no time to do the this conversion as shown in the following figure, you could give me some materials that is helpful to implement it by myself. The interpretation of a black box model' s interaction behaviors in the non-probability space is less intutive than that in the probability space. Thank you!

Can you let me know why I get the following error when I use model_output="probability" in TreeExplainer? ValueError: Only model_output="raw" is supported for feature_perturbation="tree_path_dependent"