approximate_predict generates new clusters when using cluster_selection_epsilon
I am clustering geolocation data, and I noticed that when I try to predict clusters on new data I get more clusters than I saw in the original training data.
Here is how I fit the data:
hdb = hdbscan.HDBSCAN(min_cluster_size=2, min_samples=1, metric='haversine',
cluster_selection_epsilon=epsilon, cluster_selection_method='eom',
prediction_data=True).fit(radian_locs)
This gives me 45 clusters. However, when I call:
pred_clusters, conf = hdbscan.approximate_predict(hdb, test_radian_locs)
I get a total of 232 clusters. I noticed that if I remove the cluster_selection_epsilon argument, the number of clusters match. Is there a way to preserve this in the approximate_predict function?
It is quite possible that these two features don't actually interact properly. I believe the approximate_predict is just using the clusters that would have been selected with the cluster_selection_epsilon argument, and thus produces different clusters.
Looking through the source code it seems to be that way; approximate_predict doesn't use the get_clusters method in _hdbscan_tree.pyx, which is what seems to do the cluster merging. Do you think it would be straightforward/useful to implement the same behavior for approximate_predict? I think for now I can try a simple KNN classifier to predict new cluster labels.
It may be a little tricky, but is probably feasible. I think for now the KNN classifier is more likely to the the useful approach. Sorry.
Just for future reference in case someone decides to tackle this issue: https://github.com/scikit-learn-contrib/hdbscan/issues/360 seems to be related to this bug.
Have a look at PR #398 for one solution to this issue.
I'm also fasing the same issue. I use several number of cluster_selection_epsilon in approximate_predict but the prediction results provide similar results for all number as we can see below:
import hdbscan
import pickle
import numpy as np
for eps in [0.005,0.01,0.1,0.5,1,2,5,10]:
name='hdbscan_'+str(eps)+'km.sav'
clusterer = pickle.load(open(name,'rb'))
labels,strengths = hdbscan.approximate_predict(clusterer, np.radians(coordinate.loc[:5,['longitude','latitude']]))
print(eps,labels,clusterer.cluster_selection_epsilon, clusterer.labels_.max())
0.005 [ 963 6229 6073 4376 4887 -1] 7.848050688613082e-07 8213 0.01 [ 965 6229 6073 4372 4887 -1] 1.5696101377226163e-06 8211 0.1 [ 965 6229 6073 4372 4887 -1] 1.5696101377226163e-05 8009 0.5 [ 965 6229 6073 4372 4887 -1] 7.848050688613081e-05 4952 1 [ 965 6229 6073 4372 4887 -1] 0.00015696101377226163 3340 2 [ 963 6229 6073 4376 4887 -1] 0.00031392202754452326 1557 5 [ 965 6229 6073 4372 4887 -1] 0.0007848050688613081 444 10 [ 965 6229 6073 4372 4887 -1] 0.0015696101377226162 160
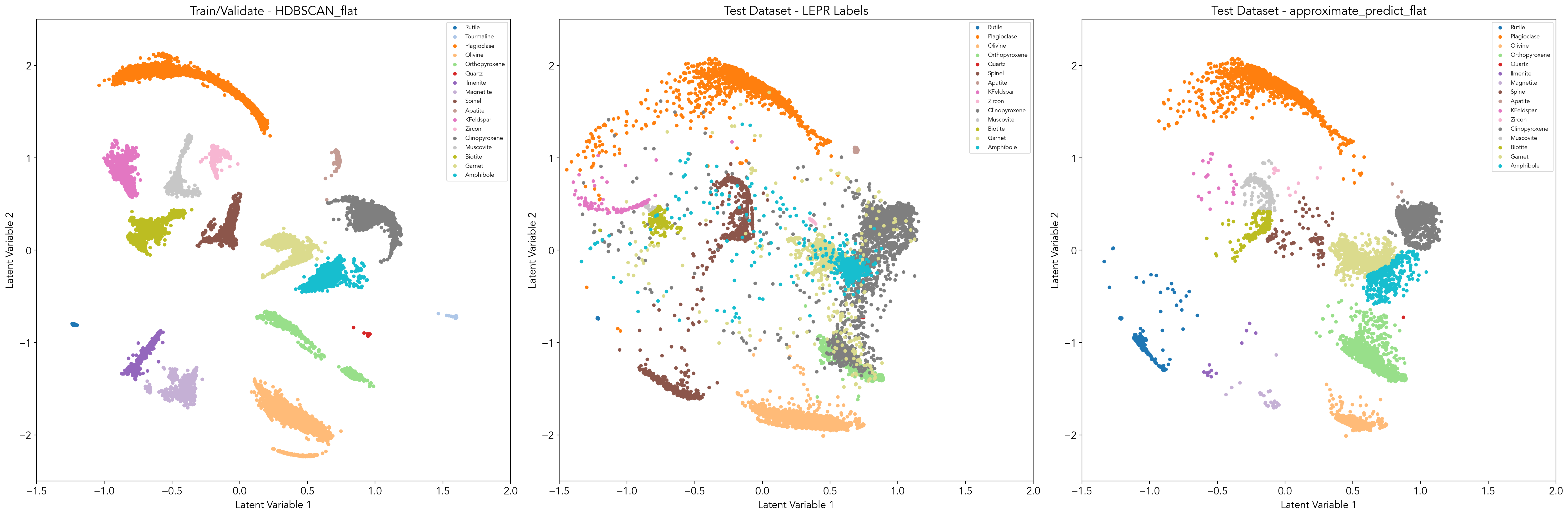
I am seeing the same issue at the moment. My latent space is currently composed of dens clusters with irregular shapes, and I've found that HDBSCAN clusters the data well when I apply the cluster_selection_epsilon. See attached for an example of this, with my test/train data on the left and the new data on the right. Most values are predicted to belong to these errant value clusters.

See the predicted labels for the test/train data (left) and new data (right) when I remove the cluster_selection_epsilon parameter. I've done quite a bit of hyperparameter tuning but can't seem to improve things too much more. I would greatly appreciate any advice for better approaches to this problem.

#398 solved this issue with the ability to select for cluster_selection_epsilon. I had to filter out all the -1 predictions, which solved this problem. See attached for updated clustering with HDBSCAN_flat and predictions from approximate_predict_flat (with the same cluster_selection_epsilon value).