Integrate SAScore as a descriptor
What does this implement/fix? Explain your changes.
This PR adds the SAScore as a proper descriptor to the RDKit as opposed to it being just a contrib. The SAScore is very popular, but calculating it accurately requires keeping the library of fragment/feature score contributions up to date. This was rightfully remarked by Peter Ertl at ICCS 2022. I've written a class to store and calculate these contributions, as well as re-implemented in C++ the code of the Python sascorer.py. I believe the way in which the contributions are calculated is correct (I emailed Peter Ertl about it some while back).
I also added the possibility of returning all individual SAScore terms. I find this helps with understanding why the values may be good/bad.
I've seen some interest in this before (https://github.com/rdkit/rdkit/issues/4161) and it got Greg’s support. That issue also mentions old fingerprint bugs affecting the SAScore output, which is concerning. I’ve also taken the chance to address issue https://github.com/rdkit/rdkit/issues/4650.
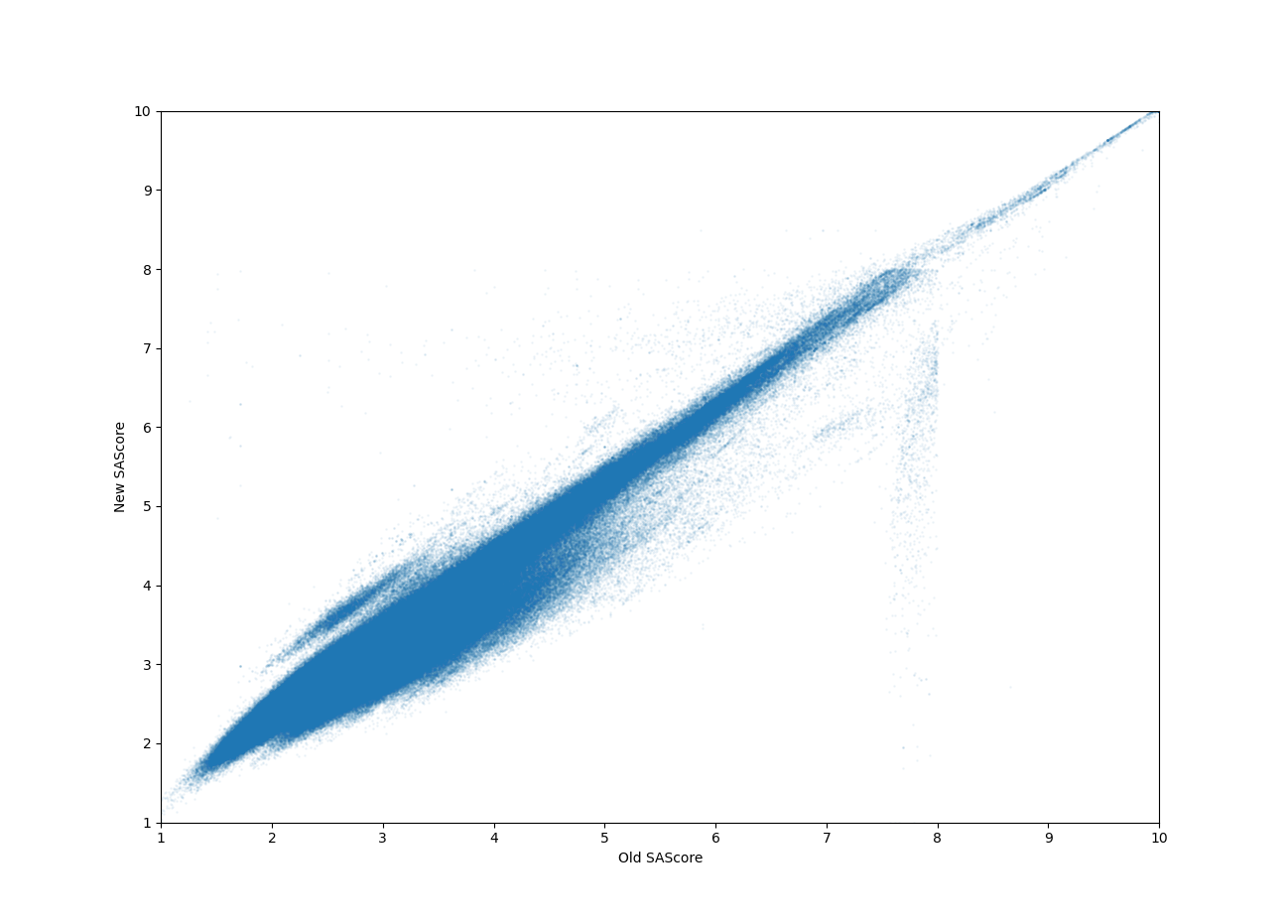
I did some preliminary tests calculating feature contributions using ChEMBL31 (small molecule drugs, no natural products, only main fragment of salts) and calculating the SAScores of the same set of molecules using both the existing Python implementation and the new C++ implementation. The values match quite well, but there is a concerning patch of molecules around OldSAScore = 8 where values are significantly off. My gut reaction was that this is related to scaling the SAScore to the 1-10 range, but I believe I’m scaling the same way as in the old implementation.
Any other comments?
This PR makes use of C++17 and Boost.Serialization to store the feature contributions. Please let me know if these are acceptable dependencies.
I’m also not sure how to implement tests for this. What should be the baseline truth?
@AlanKerstjens I like the idea of doing this, and thanks for getting started with it.
To answer your questions: using C++17 features is perfectly fine
Figuring out how to store the feature contributions is a bit trickier. It's clearly ok to use boost::serialization (probably combined with boost::iostreams so that the contributions can be stored compressed... the file is otherwise fairly large), but we'll need to make sure that the descriptor is only available when RDK_USE_BOOST_SERIALIZATION (and RDK_USE_BOOST_IOSTREAMS) is enabled and we'll have to figure out how to package the file (where to put it) so that it shows up for the various packaging schemes.
It would be better if we could directly include the feature contributions in a source file (so that no packaging is required), but I think that's going to lead to an unmanageably large file: the current version of the parameters has >705K weights
If we can't directly incorporate it into the source, having the binary (and compressed) data file is better than nothing.
It would be good to also have code available which makes it easy to update/generate the feature contributions based upon some text output. This would allow us to keep those up-to-date as RDKit versions change.
sorry, clicked the wrong button, I didn't mean to close this.
This PR was marked as stale because it has been open for 60 days with no activity.
This PR was closed because it has been inactive for 14 days since being marked as stale.