rdkit
 rdkit copied to clipboard
rdkit copied to clipboard
RGD with tetrazole core yields to core that cannot be kekulized
To Reproduce
from rdkit import Chem
from rdkit.Chem import rdRGroupDecomposition
core = Chem.MolFromSmiles("n1nn[nH]c1Cc1ccccc1")
core
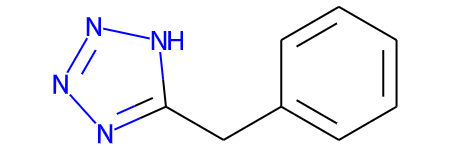
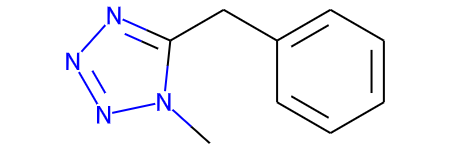
mol = Chem.MolFromSmiles("n1nnn(C)c1Cc1ccccc1")
mol
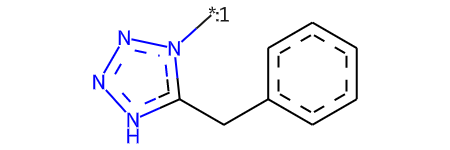
rgd = rdRGroupDecomposition.RGroupDecomposition(core)
rgd.Add(mol)
0
rgd.Process()
True
rgd.GetRGroupsAsRows(asSmiles=True)
[{'Core': 'c1ccc(Cc2[nH]nnn2[*:1])cc1', 'R1': 'C[*:1]'}]
Chem.MolFromSmiles('c1ccc(Cc2[nH]nnn2[*:1])cc1')
[16:31:47] Can't kekulize mol. Unkekulized atoms: 5 7 8
rgd.GetRGroupsAsRows()[0]["Core"]
Hi @ptosco, I met this problem before.
The RGD is problematic when dealing with core that contains chemical groups with aromatic nitrogens.
Our workaround on this issue is to convert the core as a SMARTS pattern, and then set the explicit hydrogen count to 0:
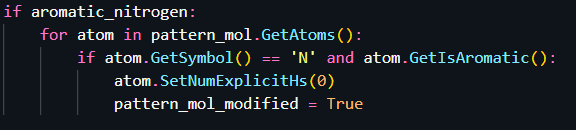
The RGD will work for this modified pattern mol. Hope this helps.
@Hong-Rui,
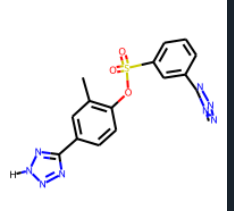
I tested doing this on this tetrazole containing molecule Cc1cc(-c2nn[nH]n2)ccc1OS(=O)(=O)c1cccc(-c2nnnn2C)c1 and then followed up with Draw.MolsToGridImage to see if it could still be drawn and I get Unkekulized atoms: 4 5 6 7 8 which are the atoms of the tetrazole ring.
You can always have rgd return the molecule version of the core and just render that. Barring that, you may have to render the smiles with sanitization off.
rgd.GetRGroupsAsRows(asSmiles=False)
Sorry I didn't mention I wasn't using RGD and I was just using the smiles as a reference to AssignBondOrdersFromTemplate. Now I realize that it can't be drawn because when you remove the hydrogen from 1H-Tetrazole with atom.SetNumExplicitHs(0) this works as a template and my actual molecule is returned correctly with the tetrazole intact with AssignBondOrdersFromTemplate (but should it? since a hydrogen of the tetrazole is now missing from reference molecule) but obviously the reference molecule cannot be drawn because the valence for the nitrogen is 2.
actual molecule before AssignBondOrdersFromTemplate and atom.SetNumExplicitHs(0) of reference smiles
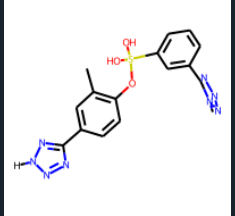
actual molecule after