Inverting stereo information when running a reaction from a SMARTS string
Describe the bug In some cases, when running a Reaction created from a SMARTS string, the stereo information is inverted next to the reactive center while it should be retained.
To Reproduce See below a simple way to reproduce the issue. Two reactions have been defined, one failing, one working. The difference between the two is only that the methanol moiety has an explicit H on the failing one.
from rdkit import Chem
from rdkit.Chem.rdChemReactions import SanitizeRxn, PreprocessReaction, ReactionFromSmarts, SanitizeFlags
from rdkit.Chem.rdmolops import AdjustQueryParameters
from rdkit.Chem.Draw import IPythonConsole
IPythonConsole.drawOptions.addAtomIndices = False
IPythonConsole.drawOptions.addStereoAnnotation = True
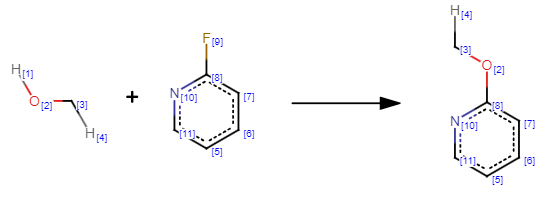
workingReaction = ReactionFromSmarts("[H:1][#8:2]-[#6:3].[F:9][c:8]1[c:7][c:6][c:5][c:11][n:10]1>>[#6:3]-[#8:2]-[c:8]1[c:7][c:6][c:5][c:11][n:10]1")
failingReaction = ReactionFromSmarts("[H:4][#6:3]-[#8:2][H:1].[F:9][c:8]1[c:7][c:6][c:5][c:11][n:10]1>>[H:4][#6:3]-[#8:2]-[c:8]1[c:7][c:6][c:5][c:11][n:10]1")
reaction = workingReaction
SanitizeRxn(reaction, sanitizeOps=SanitizeFlags.SANITIZE_ALL)
reaction.Initialize()
n_warnings, n_errors, n_reactants, n_products, labels = PreprocessReaction(reaction)
print(n_warnings, n_errors, n_reactants, n_products, labels)
for reactant in reaction.GetReactants():
reactant.UpdatePropertyCache(False)
Chem.SetAromaticity(reactant)
reactant1 = Chem.MolFromSmiles('O=C1C[C@H](O)CCN1') # (R) stereocenter
reactant2 = Chem.MolFromSmiles('CC(C)c1ccc(F)nc1C')
print(reaction.IsMoleculeReactant(reactant1)) # True
print(reaction.IsMoleculeReactant(reactant2)) # True
try:
products = reaction.RunReactants((reactant1, reactant2))
except Exception as e:
print(e)
prod = products[0][0]
Chem.SanitizeMol(prod)
print(Chem.MolToSmiles(prod))
# workingReaction (R) stereocenter => Cc1nc(O[C@@H]2CCNC(=O)C2)ccc1C(C)
# failingReaction (S) stereocenter => [H][C@]1(Oc2ccc(C(C)C)c(C)n2)CCNC(=O)C1
Chem.Draw.MolsToGridImage((prod, reactant1), subImgSize=(400,250))
Expected behavior Given my reactant matches the reaction I would expect the stereochemistry to be retained.
Screenshots
Working Reaction and output

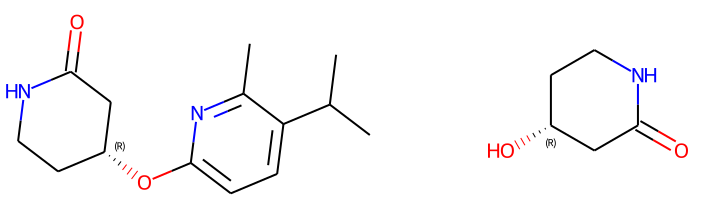
Failing reaction and output
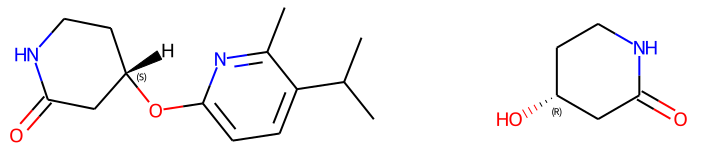
Configuration (please complete the following information):
- RDKit version: 2022.03.3
- OS: Ubuntu 18-04
- Python version (if relevant):3.8.12
- Are you using conda? yes
- If you are using conda, which channel did you install the rdkit from? conda-forge
My impression is you are doing something wrong:
reaction = AllChem.ReactionFromSmarts("[H:4][#6:3]-[O:2][#1:21].[F:9][c:8]1[c:7][c:6][c:5][c:11][n:10]1>>[H:4][#6:3]-[O:2]-[c:8]1[c:7][c:6][c:5][c:11][n:10]1.[F:9][#1:21]")
reaction.Initialize()
reactant1 = Chem.MolFromSmiles('[H][C@@]1(O)CCNC(=O)C1')
reactant2 = Chem.MolFromSmiles('CC(C)c1ccc(F)nc1C')
products = reaction.RunReactants((AllChem.AddHs(reactant1), reactant2))
print(Chem.MolToSmiles(AllChem.RemoveHs(products[0][0])))
Cc1nc(O[C@@H]2CCNC(=O)C2)ccc1C(C)C
Hi @wopozka,
Thank you for your input. I tried with your suggestion of Adding and Removing Hs and indeed it seems to help.
However, it seems incompatible with the SanitizeRxn(reaction, sanitizeOps=SanitizeFlags.SANITIZE_ALL). If I keep this line, I have an error with a pentavalent C (the stereogenic one). If I remove it, all works fine.
Digging a bit more, it seems the error only happens if I use the SanitizeFlags.SANITIZE_MERGEHS flag. I couldn't find exactly what it does. It seem to also aromatize molecules, but it is not relevant for my reactant here. Do you have any insight?
@MaxDNG It is not incompatible, it is just wrong reaction SMARTS you are using. During handling the reaction some of hydrogens are copied, and if you do not handle it properly (in reaction SMARTS) then are copied to products, and that results in sanitization error. Please proviede the simplest example you meet this problem and then I can comment.
I see. Actually the reaction SMARTS was generated by MarvinJS. Do you mean we need to sanitize it and remove extraneous Hs before?
As a matter of fact, if I use this reaction: ReactionFromSmarts("[H:3][#6:1]-[#8:2][H:6].[#6:4][F:5]>>[#6:1]-[#8:2]-[#6:4]"), it works even when I use the SanitizeFlags.SANITIZE_ALL.
@MaxDNG, Using Smarts from Marvin is not good idea, it is better to create smarts by yourself. As I said, until you show me reaction SMARTS which results in broken molecule (sanitization failed) I am not able to explain why. It is in most cases SMARTS error, in the way, that you do not cover all atoms, and some of them are just copied to final molecule, which results in zanitization error.