dask-cuda
 dask-cuda copied to clipboard
dask-cuda copied to clipboard
Test Pack/Unpack w/ cuDF Merge
@charlesbluca recently added a new serialization method for cuDF Dataframes where data is now serialized into two buffers: one for metadata and another for the data itself. As opposed to a frame per column which is currently done now
We think this may have an impact on performance when communicating and when spilling as both require serialization. I would expect the serializing time to be somewhat similar but when communicating or when spilling we iterate through all the buffers and this can negatively impact performance. Having one large buffer, we think, should improve both comms and spilling.
However, serializing with one large buffer may lead to more memory pressure as because we need to allocate a large buffer so we shouldn't be surprised if we need to tune device spilling.
Running the cuDF Merge Benchmark with pack/unpack serialization enabled would be a good place to start in trying to understand performance and tradeoffs with memory.
Ran the benchmarks with a couple different chunk sizes / device memory limits set, here are some plots of the resulting throughputs:
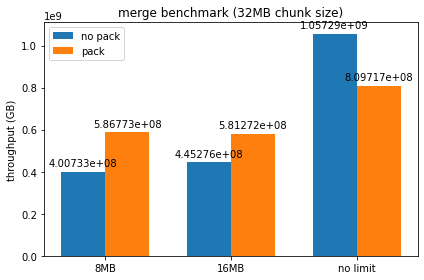
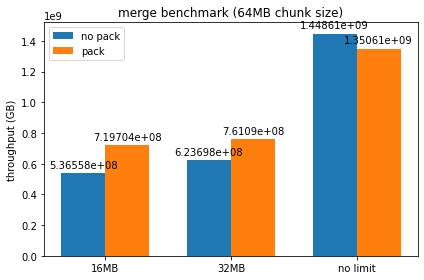
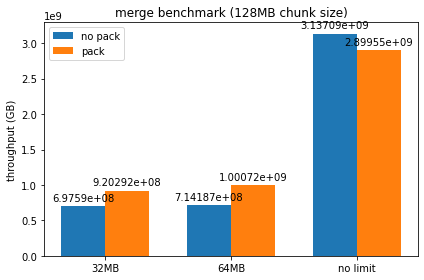

It looks like packed serialization does worse than normal serialization for smaller chunk sizes until we force spilling to host, though as chunk size gets larger it begins to perform comparably. I can try out larger chunk sizes, but at a certain threshold I begin to get a lot of RMM failures due to max pool size being exceeded when using packed serialization. I'm not entirely sure if this is just the packed serialization at fault, as it seems consistently tied to the number of runs being done by the benchmark - should we be freeing up the persisted memory on each run, or do we want all the dataframes to be persisted by design?
For context, the script used to generate these results:
for size in 1000000 2000000 4000000 8000000
do
for frac in 1 2 4
do
if [[ $frac -eq 1 ]]
then
DASK_JIT_UNSPILL=1 UCX_MAX_RNDV_RAILS=1 UCX_MEMTYPE_REG_WHOLE_ALLOC_TYPES=cuda python local_cudf_merge.py -d 0,1,2,3 -p ucx --enable-tcp-over-ucx --enable-infiniband --enable-nvlink --enable-rdmacm --interface ib0 -c $size --benchmark-json rows_"$size"_no_limit --runs 5
else
let "limit = ($size * 32) / $frac"
DASK_JIT_UNSPILL=1 UCX_MAX_RNDV_RAILS=1 UCX_MEMTYPE_REG_WHOLE_ALLOC_TYPES=cuda python local_cudf_merge.py -d 0,1,2,3 -p ucx --enable-tcp-over-ucx --enable-infiniband --enable-nvlink --enable-rdmacm --interface ib0 -c $size --device-memory-limit $limit --benchmark-json rows_"$size"_limit_"$limit"_bytes --runs 5
fi
done
done
or do we want all the dataframes to be persisted by design?
This is by design, as we want to benchmark only the compute phase. However, if generating new data for each run is important for the benchmark you're doing, I suggest adding a new flag that will force freeing data and generating again, perhaps something like --no-persist.
Is there some way to measure the cost of allocating & copying into packed form separately? Also is that included in the bandwidth measurements above or is that excluded?
if generating new data for each run is important for the benchmark you're doing
From the below lines, I was under the impression that new data was being generated for each run, unless I'm misunderstanding something about how get_random_ddf() works:
https://github.com/rapidsai/dask-cuda/blob/a0217f45e78d8f786dd0f631e38c0d9b310fb6d2/dask_cuda/benchmarks/local_cudf_merge.py#L155-L160
My concern is (assuming we are generating new data per run) that we are keeping redundant dataframes in memory after each run, which will eventually cause the GPU to run out of memory regardless of chunk size. Should we be deleting these dataframes from memory after each run?
Is there some way to measure the cost of allocating & copying into packed form separately? Also is that included in the bandwidth measurements above or is that excluded?
I'd imagine this is something that needs to happen at the cuDF level? We could probably track the time it takes to perform a single dataframe packed serialization/deserialization operation and use that to get throughput of packed to unpacked memory. This cost is included in the bandwidth measurements above - would we be interested in calculating these bandwidth measurements with this cost excluded? That is, only compute the time it takes to send a PackedColumns over the wire, but not the time it takes to pack/unpack it.
Is there some way to measure the cost of allocating & copying into packed form separately? Also is that included in the bandwidth measurements above or is that excluded?
I'd imagine this is something that needs to happen at the cuDF level? We could probably track the time it takes to perform a single dataframe packed serialization/deserialization operation and use that to get throughput of packed to unpacked memory. This cost is included in the bandwidth measurements above - would we be interested in calculating these bandwidth measurements with this cost excluded? That is, only compute the time it takes to send a PackedColumns over the wire, but not the time it takes to pack/unpack it.
Yeah I think so. More generally it would be good to disentangle these things somehow (even if that is just timing the packing alone). This will likely improve our intuitions about when packing makes sense.
It could also motivate different efforts. IOW if we know transmission is sped up X times (where this is significant), but we pay a cost of Y for packing, then we can discuss with others (particularly on the memory allocation/copying side of things) how to cutdown Y.
A second thing worth looking at is different #s of columns for the same size of data. IOW how does the degree of fragmentation play into whether we should choose to pack or not? Naively would guess increased fragmentation means packing is more valuable, but I could be wrong about this.
I added some NVTX annotations for the serialization to check out what's happening there - we definitely spend a lot of time on the libcudf pack() operation, but it also looks like in some cases we spend significantly more time on the index equality check we do to assess if we want to drop the index or not. I think this is because of a cuModuleLoadData call, which I imagine needs to be called the first time binary_operation is called (using ~256MB chunks, no device limit here):
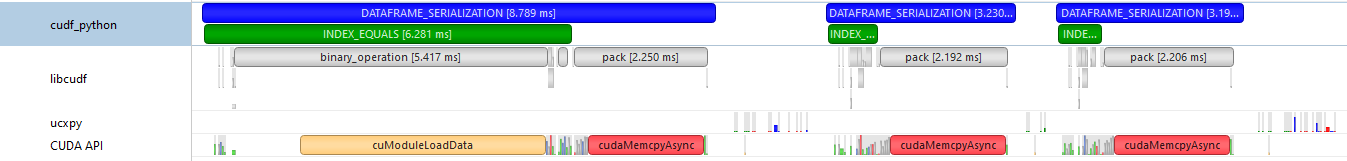
Maybe for the sake of performance it would be better not to do this check? It is only there to prevent us from packing a RangeIndex that can be reconstructed on the fly, so I'm not sure what the memory implications would be here for larger dataframes.
On average, it looks like serialization without packing is around 30x faster than with, though this is magnified for the longer durations:


It looks like the longest packed serialization durations aren't actually related to the long INDEX_EQUALS call, but longer pack() calls:

Based on the duration here, I would guess that this operation is serializing the merged dataframe? Though I don't know enough about how merge works in Dask to say this with certainty.
I'm interested in profiling these same commands with a lower device memory limit, since that showed gains from using packed serialization, but I think I might focus now on just timing basic pack operations to get a sense of how dataframe size / fragmentation impacts performance.
Maybe for the sake of performance it would be better not to do this check?
Found a much faster way to perform this check without doing an element-by-element comparison rapidsai/cudf#8917:

Looks like there was memory leaking happening with PackedColumns that should be fixed by rapidsai/cudf#8936. Once that is in, interested in if that will have noticeable performance implications for larger chunk sizes
Thanks for following up @charlesbluca
Timed a few runs of packing dataframes of int64s with different sizes/fragmentation, here's a plot of the average throughput:
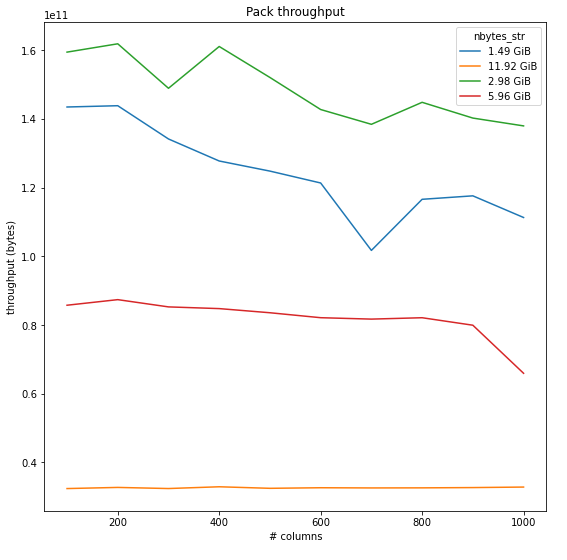
From this, it looks like we would see the highest unpacked -> packed throughput on dataframes around 2-5 GiB; it also seems like it generally goes down as fragmentation increases, though I haven't checked for very small numbers of columns. For reference, the average times for the pack operation:

Packed -> unpacked memory generally performed the same regardless of size, with decreased throughput as fragmentation went up:

I'm looking into making nvcomp available in cudf and/or dask-cudf. It sounds like you want to be able to apply compression to the packed dataframes, then when a computation is needed on a portion of the dataframe, only the portion that is needed is unpacked and decompressed. Is that right?
@thomcom am going to send you some more info offline. Happy to chat more if you are interested 🙂
Also related to the nvcomp discussion is issue ( https://github.com/rapidsai/dask-cuda/issues/760 )
This issue has been labeled inactive-30d due to no recent activity in the past 30 days. Please close this issue if no further response or action is needed. Otherwise, please respond with a comment indicating any updates or changes to the original issue and/or confirm this issue still needs to be addressed. This issue will be labeled inactive-90d if there is no activity in the next 60 days.
This issue has been labeled inactive-90d due to no recent activity in the past 90 days. Please close this issue if no further response or action is needed. Otherwise, please respond with a comment indicating any updates or changes to the original issue and/or confirm this issue still needs to be addressed.
This issue has been labeled inactive-30d due to no recent activity in the past 30 days. Please close this issue if no further response or action is needed. Otherwise, please respond with a comment indicating any updates or changes to the original issue and/or confirm this issue still needs to be addressed. This issue will be labeled inactive-90d if there is no activity in the next 60 days.
This issue has been labeled inactive-90d due to no recent activity in the past 90 days. Please close this issue if no further response or action is needed. Otherwise, please respond with a comment indicating any updates or changes to the original issue and/or confirm this issue still needs to be addressed.