odbc
 odbc copied to clipboard
odbc copied to clipboard
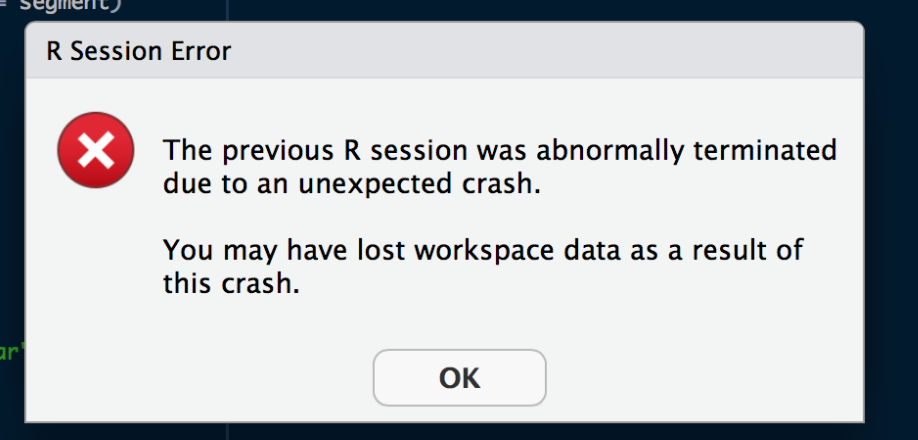
Previewing tables via the connections pane in R Studio Server leads to an "R Session Error"
Issue Description and Expected Result
Previewing tables via the connections pane in R Studio Server leads to an "R session error":
Within the connections pane...
...pressing the icon on the right (to preview the table)...

...leads to this for about a minute...

...and then this message...
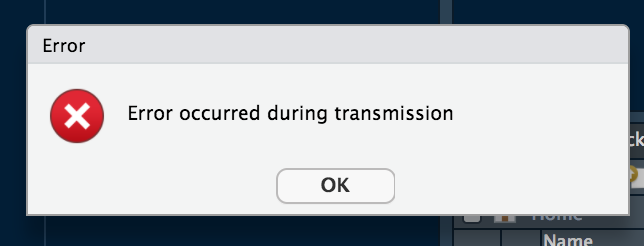
...followed by this message, and no preview of the table.
We expected to see a new tab opened with the first 1,000 rows of the table.
We have experienced this issue repeatedly and have been unable to successfully preview many tables from our database. One example of such a table has 121MM rows, 15 columns (numbers, 1 string, a few dates). Another example of such a table has 104MM rows, 16 columns (integer, numeric, timestamp, character).
One table from the same database that we were able to preview successfully has 902,000 rows, 9 columns (character, timestamp, integer)
We are able to successfully pull the first 1,000 rows from all tables in an R script, but for some tables we are unable to preview through the connections pane.
Please let us know if any additional detail will help in resolving this issue.
Thank you.
Database
Amazon Redshift Version: 8.0.2
Reproducible Example
Session Info
#session info from before crash
devtools::session_info()
#> output
Session info --------------------------------------------------------------------------------------
setting value
version R version 3.4.2 (2017-09-28)
system x86_64, linux-gnu
ui RStudio (1.1.383)
language (EN)
collate en_US.UTF-8
tz Etc/UTC
date 2017-12-13
Packages ----------------------------------------------------------------------------------------------------------
package * version date source
base * 3.4.2 2017-10-27 local
bit 1.1-12 2014-04-09 cran (@1.1-12)
bit64 0.9-7 2017-05-08 cran (@0.9-7)
blob 1.1.0 2017-06-17 cran (@1.1.0)
compiler 3.4.2 2017-10-27 local
datasets * 3.4.2 2017-10-27 local
DBI 0.7 2017-06-18 cran (@0.7)
devtools 1.13.4 2017-11-09 CRAN (R 3.4.2)
digest 0.6.12 2017-01-27 CRAN (R 3.4.2)
graphics * 3.4.2 2017-10-27 local
grDevices * 3.4.2 2017-10-27 local
hms 0.3 2016-11-22 cran (@0.3)
memoise 1.1.0 2017-04-21 CRAN (R 3.4.2)
methods * 3.4.2 2017-10-27 local
odbc * 1.1.3.9000 2017-11-06 Github (rstats-db/odbc@b3d159a)
pillar 0.0.0.9000 2017-11-09 Github (r-lib/pillar@99ef1d8)
Rcpp 0.12.14 2017-11-23 cran (@0.12.14)
rlang 0.1.4.9000 2017-12-12 Github (tidyverse/rlang@5ebcf24)
rstudioapi 0.7 2017-09-07 CRAN (R 3.4.2)
stats * 3.4.2 2017-10-27 local
tibble 1.3.4.9002 2017-11-09 Github (tidyverse/tibble@1ae69ee)
tools 3.4.2 2017-10-27 local
utils * 3.4.2 2017-10-27 local
withr 2.1.0 2017-11-01 CRAN (R 3.4.2)
yaml 2.1.14 2016-11-12 CRAN (R 3.4.2)
Same experience here - I am using PostgreSQL 9.3.20 on x86_64-unknown-linux-gnu, although it looks to me like the issue here is apparent in the Connections tab code:
dbGetQuery(connection, paste("SELECT * FROM", name), n = rowLimit)
To test, then (on a table with ~7M rows):
library(odbc)
con <- dbConnect(odbc::odbc(), driver = "PostgreSQL", host = "localhost", uid = "rstudio_admin",
pwd = "admin_user_be_careful", port = "5432", Database = "postgres")
system.time(odbc::dbGetQuery(con, "SELECT * FROM datawarehouse.flight LIMIT 1000"))
#> user system elapsed
#> 0.007 0.000 0.018
system.time(odbc::dbGetQuery(con, "SELECT * FROM datawarehouse.flight", n=1000))
#> fails... takes a long time... etc.
Further, we can see on the server side what is being executed:

I think this goes back to the implementation of dbGetQuery, which uses n to limit the dbFetch, not the query itself. On Postgres/Redshift, this seems to equate to executing the entire query and preparing the result set, but then only returning the first 1000 rows. Memory issues / size issues / etc. result.
setMethod("dbGetQuery", signature("OdbcConnection", "character"),
function(conn, statement, n = -1, ...) {
rs <- dbSendQuery(conn, statement, ...)
on.exit(dbClearResult(rs))
df <- dbFetch(rs, n = n, ...)
if (!dbHasCompleted(rs)) {
warning("Pending rows", call. = FALSE)
}
df
}
)
Unfortunately there is no good universal way to limit the size of a query that works across all databases. What you can do is define a database specific version of dbGetQuery for PostgreSQL or Redshift which uses LIMIT to restrict the query. e.g.
setMethod("dbGetQuery", signature("Redshift", "character"),
function(conn, statement, n = -1, ...) {
if (n > -1) {
statement <- paste(statement, "LIMIT ", n)
}
rs <- dbSendQuery(conn, statement, ...)
on.exit(dbClearResult(rs))
df <- dbFetch(rs, n = n, ...)
if (!dbHasCompleted(rs)) {
warning("Pending rows", call. = FALSE)
}
df
})
Is this something you'd be willing to accept a PR for, @jimhester? I know it clutters up the code-base with database specific shenanigans. Anything else we can assist with?
I think this would be best as a new generic in DBI, e.g. DBI::dbPreviewQuery() with default method that wrapped the query in a subquery with a limit clause.
I like that. FWIW, I recall ChartIO having similar behavior. It would yell at you if you added a semicolon or put a LIMIT statement in your query, since those are pretty common things that would mess up the dbPreviewQuery code. The perk is that this would be most useful for the connections pane, and we don't have to worry about a user-provided query there. Happy to contribute where possible!
Oh in that case the generic should be dbPreviewTable()
Maybe add an (optional) n argument to dbReadTable() ?
We now have odbcPreviewQuery() so this should be straightforward.