satpy
 satpy copied to clipboard
satpy copied to clipboard
MSG-SEVIRI L1 (native): reader_kwargs 'calib_mode':'GSICS' leads to dataset being only filled with NaNs (e.g. IR_108)
Describe the bug
When opening several MSG-SEVIRI-Level1b files in the native format, I encountered the problem that when I define the "calib_mode" to be "GSICS" in some cases the data of channels I wanted to work with were all NaN (e.g. IR_108, IR_120).
If the calibration is set to "NOMINAL", the data are available.
Until now it only occured with MSG-SEVIRI data on 07.05.2016 from 00:57:42 UTC onwards. (I only checked unil the 08.05.2016 00:00:00 UTC)
There are no error messages, only when plotting or trying to work with the GSICS-calibrated data the warning comes that the array only consists of NaNs.
It appears to be related with the information in the header of the file. For the file shown in the code (MSG3-SEVI-MSG15-0100-NA-20160507101241.351000000Z-NA.nat) the header contains the following values for the GSICS coefficients:
- 'GSICSCalCoeff': array([ 0., 0., 0., inf, inf, inf, inf, inf, inf, inf, inf, 0.],dtype=float32),
- 'GSICSCalError': array([ 0., 0., 0., nan, nan, nan, nan, nan, nan, nan, nan, 0.],dtype=float32),
- 'GSICSOffsetCount': array([-51., -51., -51., -51., -51., -51., -51., -51., -51., -51., -51., -51.],dtype=float32)
In other files these have actual values instead of the inf and nans. (e.g. MSG3-SEVI-MSG15-0100-NA-20171013001239.960000000Z-NA.nat):
- 'GSICSCalCoeff': array([0. , 0. , 0. , 0.00361306, 0.00837507, 0.03874956, 0.12729841, 0.10432485, 0.20525225, 0.2221154 , 0.16503815, 0. ], dtype=float32),
- 'GSICSCalError': array([0.0000000e+00, 0.0000000e+00, 0.0000000e+00, 3.1434915e-06, 6.5775525e-06, 2.9304914e-05, 8.3134779e-05, 9.6331736e-05, 1.3235498e-04, 1.7277416e-04, 2.3843742e-04, .0000000e+00], dtype=float32),
- 'GSICSOffsetCount': array([-51. , -51. , -51. , -51.278328, -50.9026 , -53.086205, -52.434498, -52.85853 , -51.441326, -50.890816, -64.03467 , -51. ], dtype=float32)
In most other files without actual values, there are only 0s for GSICSCalCoeff and GSICSCalError and -51s for GSICSOffsetCount. (e.g. MSG2-SEVI-MSG15-0100-NA-20100601232742.293000000Z-NA.nat)
To Reproduce
import satpy
import matplotlib.pyplot as plt
msg_file="./MSG3-SEVI-MSG15-0100-NA-20160507101241.351000000Z-NA.nat" # (downloaded from data.eumesat.int)
scn_nominal=satpy.scene.Scene([msg_file],reader='seviri_l1b_native',reader_kwargs={'fill_disk':True,'calib_mode':'NOMINAL'})
scn_nominal.load(['IR_108'],calibration=['brightness_temperature'], upper_right_corner='NE')
scn_gsics=satpy.scene.Scene([msg_file],reader='seviri_l1b_native',reader_kwargs={'fill_disk':True,'calib_mode':'GSICS'})
scn_gsics.load(['IR_108'],calibration=['brightness_temperature'], upper_right_corner='NE')
plt.figure(dpi=130)
plt.title('BT of IR_108 for calib_mode: NOMINAL \n 2016-05-07 10:12:41')
plt.imshow(scn_nominal['IR_108'],cmap='RdYlBu_r')
plt.colorbar()
plt.figure(dpi=130)
plt.title('BT of IR_108 for calib_mode: GSICS \n 2016-05-07 10:12:41')
plt.imshow(scn_gsics['IR_108'],cmap='RdYlBu_r')
plt.colorbar()
Expected behavior On other dates (e.g. 01.06.2010 23:27:42 UTC, 13.10.2017 00:12:39 UTC) both NOMINAL and GSICS yield valid data fields.
Actual results No error-messages shown.
Figures
Here is the result for NOMINAL:
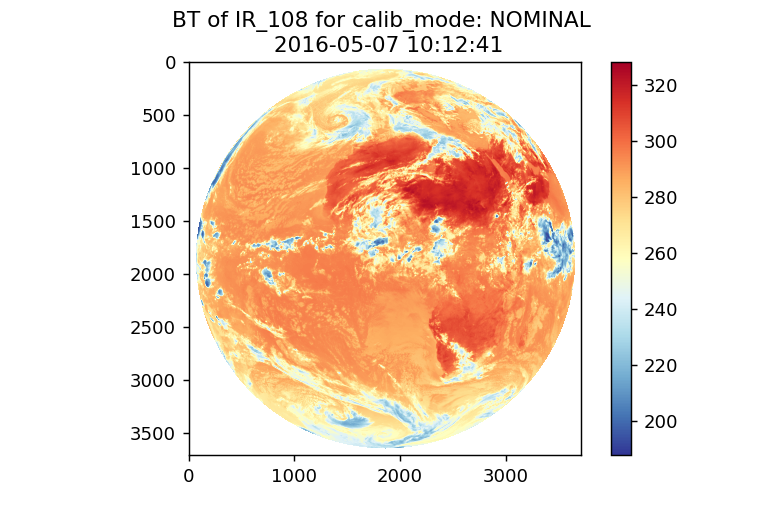
Here is the result for GSICS: (only NaNs)
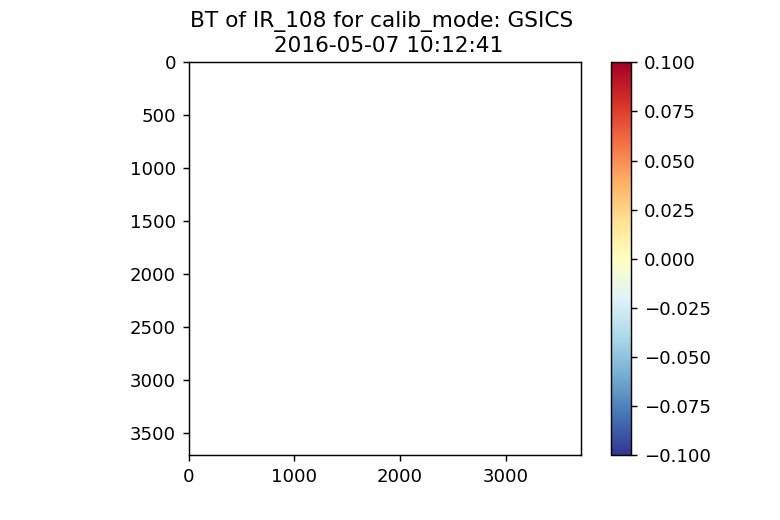
Environment Info:
- OS: Linux
- Satpy Version: 0.31.0
(This is my first issue... I hope everything is comprehensible)
This appears to be an issue with the SEVIRI data itself. But does suggest a more robust check should be made in the reader to catch cases where the raw data is bad.