botorch
 botorch copied to clipboard
botorch copied to clipboard
Weird fitting behavior after condition_on_observations
Using fit_gpytorch_model on a model obtained using condition_on_observations produces bogus fits. This was described in https://github.com/pytorch/botorch/issues/337#issuecomment-563358045.
Basic model:
import torch
from torch import Tensor
from botorch.models import SingleTaskGP
from botorch.fit import fit_gpytorch_model
from gpytorch.mlls import ExactMarginalLogLikelihood
from gpytorch.likelihoods import GaussianLikelihood
from gpytorch.constraints.constraints import GreaterThan
from gpytorch.priors.torch_priors import GammaPrior
import numpy as np
import matplotlib.pyplot as plt
from time import sleep
%matplotlib tk
tx = Tensor([[0.4963, 0.7682],
[0.0885, 0.1320],
[0.3074, 0.6341],
[0.4901, 0.8964],
[0.4556, 0.6323],
[0.3489, 0.4017],
[0.4716, 0.0649],
[0.4607, 0.1010],
[0.8607, 0.8123],
[0.8981, 0.0747],
[0.6985, 0.2838],
[0.4153, 0.2880]])
ty = Tensor([[77.3542],
[136.5441],
[27.0687],
[112.9234],
[43.0725],
[19.5122],
[10.5993],
[10.6371],
[123.6821],
[4.9352],
[26.4374],
[13.3470]])
def contour_plotter(model):
fig, ax = plt.subplots(ncols=2, figsize=(8, 4))
# fig.tight_layout()
ax[0].set_title("$\\mu_n$")
ax[1].set_title("$\\Sigma_n$")
for x in ax:
x.scatter(model.train_inputs[0].numpy()[:, 0], model.train_inputs[0].numpy()[:, 1], marker='x', color='black')
x.set_aspect('equal')
x.set_xlabel("x")
x.set_xlim(0, 1)
x.set_ylim(0, 1)
plt.show(block=False)
# plot the mu
k = 100
x = torch.linspace(0, 1, k)
xx, yy = np.meshgrid(x, x)
xy = torch.cat([Tensor(xx).unsqueeze(-1), Tensor(yy).unsqueeze(-1)], -1)
means = model.posterior(xy).mean.squeeze().detach().numpy()
c = ax[0].contourf(xx, yy, means, alpha=0.8)
plt.colorbar(c, ax=ax[0])
# plot the Sigma
x = torch.linspace(0, 1, k)
xx, yy = np.meshgrid(x, x)
xy = torch.cat([Tensor(xx).unsqueeze(-1), Tensor(yy).unsqueeze(-1)], -1)
means = model.posterior(xy).variance.pow(1 / 2).squeeze().detach().numpy()
c = ax[1].contourf(xx, yy, means, alpha=0.8)
plt.colorbar(c, ax=ax[1])
plt.show(block=False)
plt.pause(0.01)
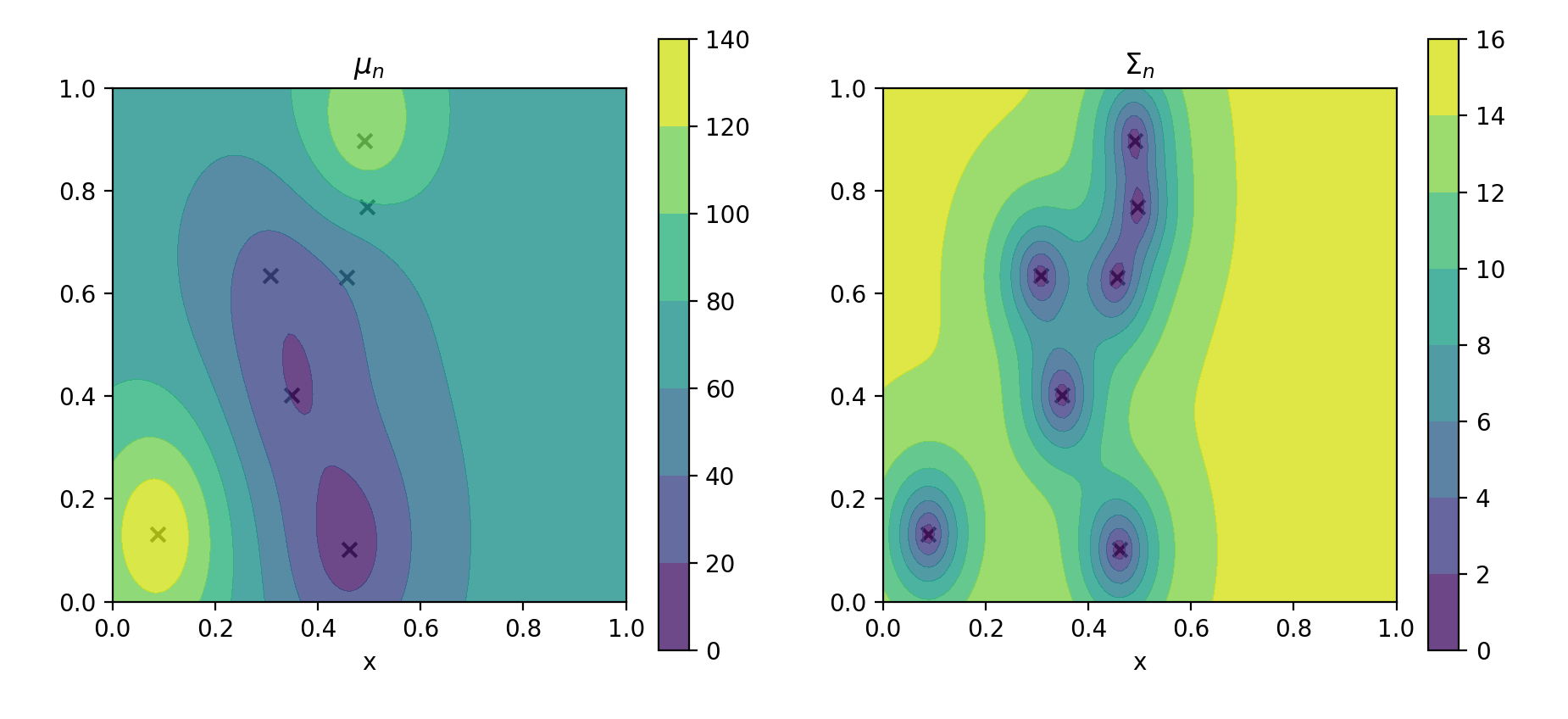
Basic model
n = 6
gp = SingleTaskGP(tx[0: n], ty[0: n], likelihood)
mll = ExactMarginalLogLikelihood(gp.likelihood, gp)
fit_gpytorch_model(mll)
contour_plotter(gp)
plt.show()
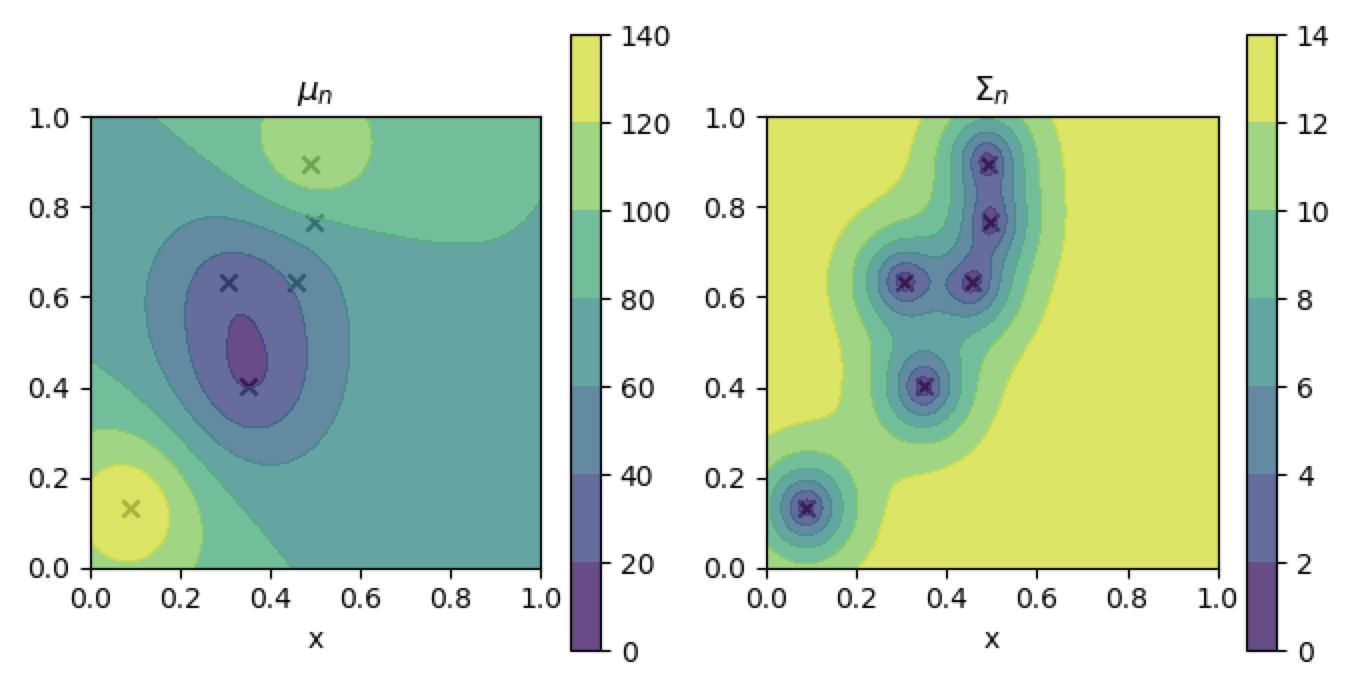
Conditioning works
candidate_point = tx[n+1, :].reshape(1, -1)
observation = ty[n+1, :].reshape(1, -1)
gp2 = gp.condition_on_observations(candidate_point, observation)
contour_plotter(gp)
plt.show()
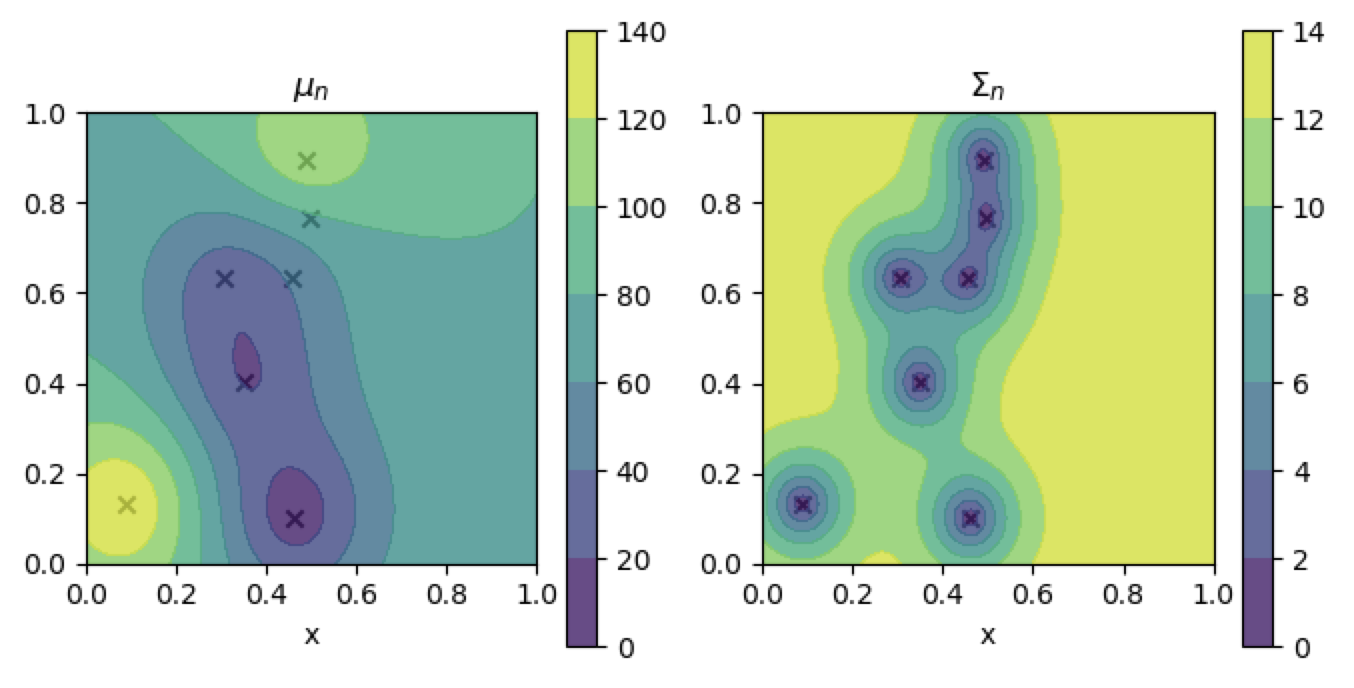
Fitting the conditioned model fails
mll2 = ExactMarginalLogLikelihood(gp2.likelihood, gp2)
fit_gpytorch_model(mll2)
contour_plotter(gp2)
plt.show()
I looked into this some more. It seems like this is caused by the closures that are used for setting/getting parameters that have a prior registered from gpytorch modules don't get properly deepcopied when the model gets deepcopied during condition_on_observations. This doesn't matter for making predictions, but messes up setting parameters / computing MLLs. cc @jacobrgardner
This seems both pretty hard to fix and a lower priority issue (since you can always warm-start your model by instantiating a new model including the additional data and subsequently loading the state dict from the previous one). Applying a wontfix for now.
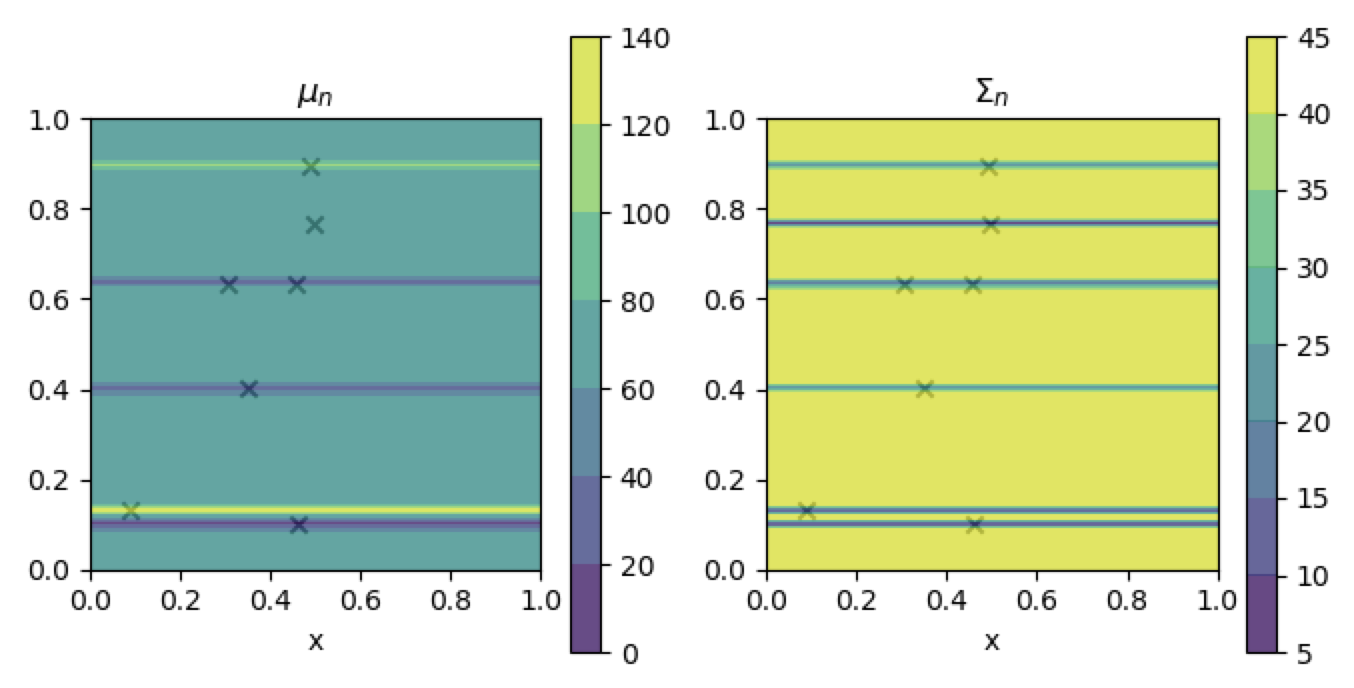
This seems to have been fixed. Here's what I'm now getting for the conditioned model: